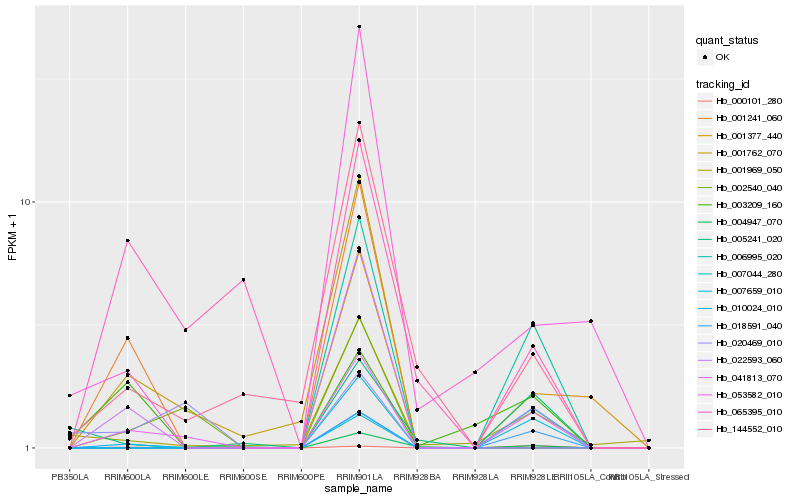
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041813_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_006995_020 |
0.2682639472 |
- |
- |
hypothetical protein glysoja_017339 [Glycine soja] |
| 3 |
Hb_018591_040 |
0.2791115014 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 4 |
Hb_007044_280 |
0.2831891138 |
- |
- |
PREDICTED: uncharacterized protein LOC105123357 [Populus euphratica] |
| 5 |
Hb_001969_050 |
0.2996229673 |
- |
- |
PREDICTED: uncharacterized protein LOC105634920 isoform X1 [Jatropha curcas] |
| 6 |
Hb_020469_010 |
0.3054564109 |
- |
- |
- |
| 7 |
Hb_144552_010 |
0.3160297064 |
- |
- |
CC-NBS-LRR protein, putative [Theobroma cacao] |
| 8 |
Hb_004947_070 |
0.3270631628 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 9 |
Hb_007659_010 |
0.3421152767 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Jatropha curcas] |
| 10 |
Hb_000101_280 |
0.3448369174 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 11 |
Hb_002540_040 |
0.3451701175 |
- |
- |
BnaC05g39550D [Brassica napus] |
| 12 |
Hb_001762_070 |
0.3466011756 |
- |
- |
- |
| 13 |
Hb_003209_160 |
0.355364045 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 14 |
Hb_065395_010 |
0.3564524621 |
- |
- |
hypothetical protein POPTR_0014s14760g, partial [Populus trichocarpa] |
| 15 |
Hb_005241_020 |
0.3584970638 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 16 |
Hb_001377_440 |
0.3590095655 |
- |
- |
- |
| 17 |
Hb_053582_010 |
0.3602329971 |
- |
- |
- |
| 18 |
Hb_001241_060 |
0.36538858 |
- |
- |
- |
| 19 |
Hb_010024_010 |
0.3655771666 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_022593_060 |
0.3668020839 |
- |
- |
- |