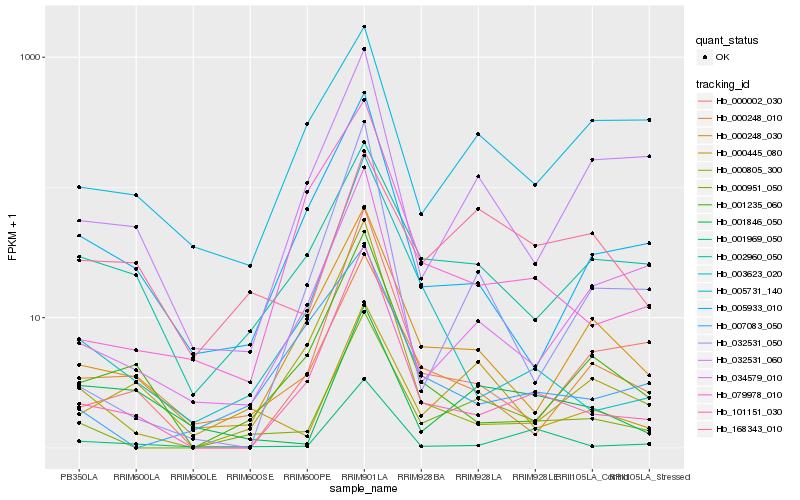
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000951_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000248_030 |
0.2230944506 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 3 |
Hb_101151_030 |
0.2287573319 |
- |
- |
- |
| 4 |
Hb_032531_050 |
0.2316125048 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 5 |
Hb_032531_060 |
0.2438605706 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 6 |
Hb_000002_030 |
0.2440045887 |
- |
- |
- |
| 7 |
Hb_002960_050 |
0.245277056 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 8 |
Hb_034579_010 |
0.2487235733 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000805_300 |
0.2514909489 |
- |
- |
- |
| 10 |
Hb_000248_010 |
0.2555697062 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 11 |
Hb_007083_050 |
0.2580204645 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 12 |
Hb_005933_010 |
0.2631004281 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 13 |
Hb_005731_140 |
0.2688375236 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 14 |
Hb_001235_060 |
0.2711720698 |
- |
- |
- |
| 15 |
Hb_001969_050 |
0.2732026258 |
- |
- |
PREDICTED: uncharacterized protein LOC105634920 isoform X1 [Jatropha curcas] |
| 16 |
Hb_168343_010 |
0.2736358795 |
- |
- |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 17 |
Hb_000445_080 |
0.2738955622 |
transcription factor |
TF Family: MIKC |
PREDICTED: floral homeotic protein PMADS 2 [Jatropha curcas] |
| 18 |
Hb_079978_010 |
0.27487217 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 19 |
Hb_003623_020 |
0.2778144436 |
- |
- |
- |
| 20 |
Hb_001846_050 |
0.2866559759 |
- |
- |
- |