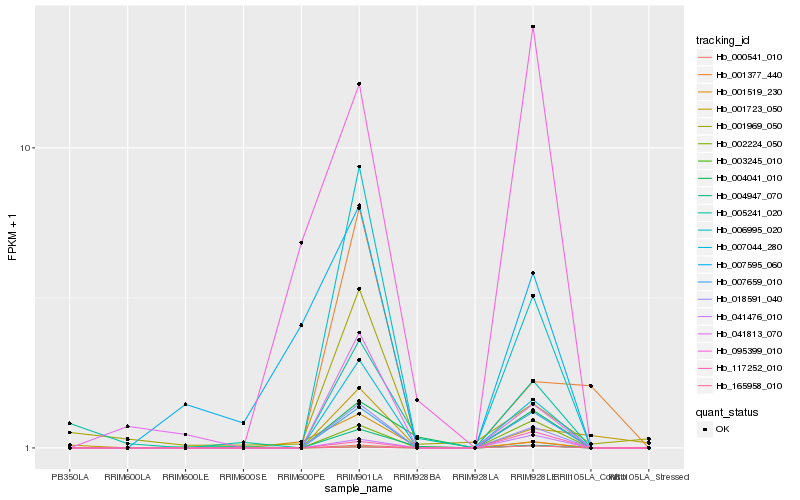
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018591_040 |
0.0 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 2 |
Hb_007044_280 |
0.0142628906 |
- |
- |
PREDICTED: uncharacterized protein LOC105123357 [Populus euphratica] |
| 3 |
Hb_006995_020 |
0.0754410414 |
- |
- |
hypothetical protein glysoja_017339 [Glycine soja] |
| 4 |
Hb_007659_010 |
0.1419021411 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Jatropha curcas] |
| 5 |
Hb_002224_050 |
0.2163419762 |
- |
- |
PREDICTED: uncharacterized protein LOC104764484 isoform X1 [Camelina sativa] |
| 6 |
Hb_041476_010 |
0.2570251078 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 7 |
Hb_004041_010 |
0.2594233409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_041813_070 |
0.2791115014 |
- |
- |
- |
| 9 |
Hb_005241_020 |
0.2893676561 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 10 |
Hb_001377_440 |
0.2953284821 |
- |
- |
- |
| 11 |
Hb_001969_050 |
0.3112407322 |
- |
- |
PREDICTED: uncharacterized protein LOC105634920 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001519_230 |
0.3174447872 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 13 |
Hb_001723_050 |
0.318370722 |
- |
- |
galactinol synthase-2 [Manihot esculenta] |
| 14 |
Hb_000541_010 |
0.3267199783 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_003245_010 |
0.3304046467 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_165958_010 |
0.3307232036 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_004947_070 |
0.3326111378 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 18 |
Hb_007595_060 |
0.3377483692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_095399_010 |
0.344253515 |
- |
- |
cytochrome b/f complex subunit IV [Sphagnum fimbriatum] |
| 20 |
Hb_117252_010 |
0.3637427298 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |