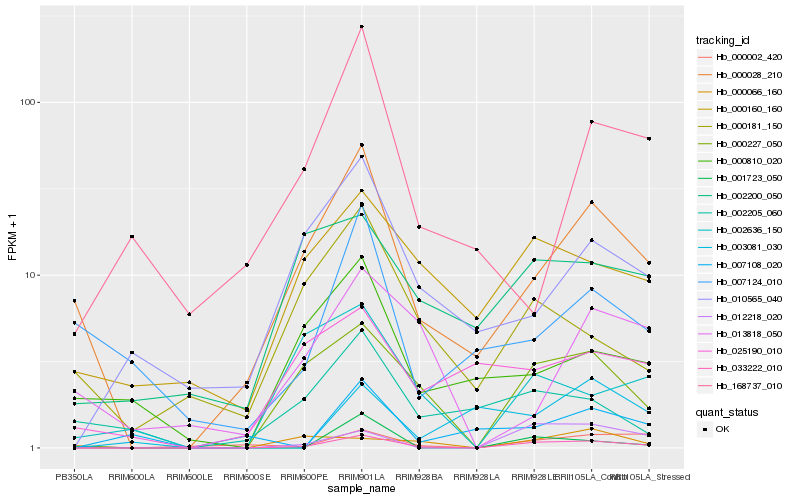
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033222_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105173350 [Sesamum indicum] |
| 2 |
Hb_000227_050 |
0.2136457421 |
- |
- |
- |
| 3 |
Hb_012218_020 |
0.2552358989 |
- |
- |
hypothetical protein JCGZ_14688 [Jatropha curcas] |
| 4 |
Hb_000028_210 |
0.2596954255 |
- |
- |
hypothetical protein CISIN_1g0103302mg, partial [Citrus sinensis] |
| 5 |
Hb_000002_420 |
0.2701192258 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1-like [Citrus sinensis] |
| 6 |
Hb_001723_050 |
0.2940464405 |
- |
- |
galactinol synthase-2 [Manihot esculenta] |
| 7 |
Hb_000160_160 |
0.3164034957 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_010565_040 |
0.326621731 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 9 |
Hb_002636_150 |
0.333728275 |
- |
- |
- |
| 10 |
Hb_002200_050 |
0.3413792494 |
- |
- |
unknown [Lotus japonicus] |
| 11 |
Hb_000181_150 |
0.3500302217 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 12 |
Hb_007108_020 |
0.3553612207 |
- |
- |
- |
| 13 |
Hb_007124_010 |
0.3566237947 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_025190_010 |
0.3568754671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_168737_010 |
0.35802806 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000810_020 |
0.3587241582 |
- |
- |
- |
| 17 |
Hb_000066_160 |
0.3591098869 |
- |
- |
- |
| 18 |
Hb_003081_030 |
0.3601992841 |
- |
- |
PREDICTED: uncharacterized protein LOC101256381 [Solanum lycopersicum] |
| 19 |
Hb_013818_050 |
0.3603269137 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002205_060 |
0.3605461418 |
- |
- |
- |