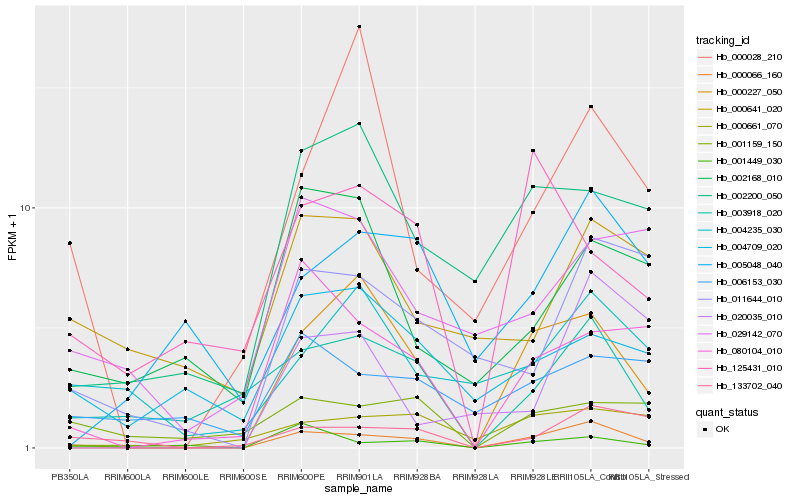
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000066_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_000227_050 |
0.2351212837 |
- |
- |
- |
| 3 |
Hb_080104_010 |
0.2646331331 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_133702_040 |
0.2659301377 |
- |
- |
- |
| 5 |
Hb_003918_020 |
0.2838008828 |
- |
- |
- |
| 6 |
Hb_011644_010 |
0.2931649679 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 7 |
Hb_000661_070 |
0.2934595481 |
- |
- |
- |
| 8 |
Hb_001449_030 |
0.3020901664 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002200_050 |
0.3051957862 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_001159_150 |
0.3101371156 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_005048_040 |
0.3158358753 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 12 |
Hb_006153_030 |
0.3185432621 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 13 |
Hb_004235_030 |
0.3190567825 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 14 |
Hb_029142_070 |
0.3212787722 |
- |
- |
- |
| 15 |
Hb_004709_020 |
0.3236599063 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 16 |
Hb_020035_010 |
0.325933191 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1-like isoform X2 [Glycine max] |
| 17 |
Hb_002168_010 |
0.3298542768 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 18 |
Hb_000641_020 |
0.3303881488 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 19 |
Hb_125431_010 |
0.3311361498 |
- |
- |
- |
| 20 |
Hb_000028_210 |
0.331821176 |
- |
- |
hypothetical protein CISIN_1g0103302mg, partial [Citrus sinensis] |