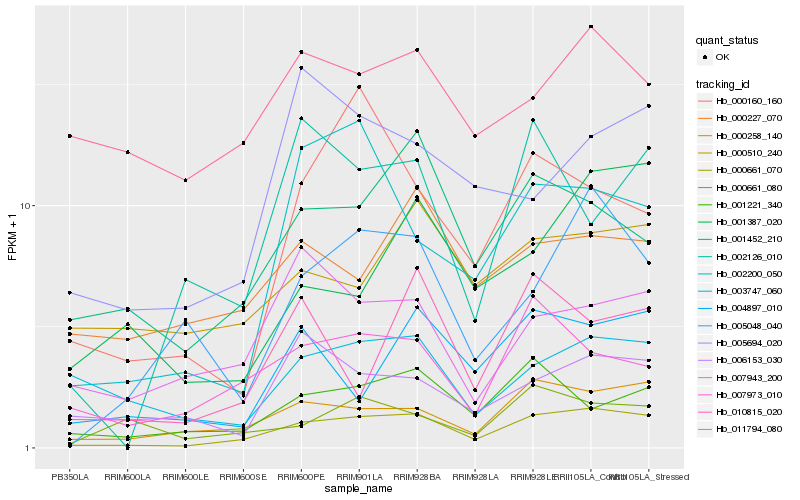
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000258_140 |
0.1513696645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001221_340 |
0.1878750148 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 4 |
Hb_007973_010 |
0.1891918917 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001452_210 |
0.1892867379 |
- |
- |
molybdopterin biosynthesis CNX1 protein [Arabidopsis thaliana] |
| 6 |
Hb_005048_040 |
0.194448236 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 7 |
Hb_004897_010 |
0.2044477623 |
- |
- |
- |
| 8 |
Hb_003747_060 |
0.2072046721 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 9 |
Hb_007943_200 |
0.2091547616 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 10 |
Hb_000510_240 |
0.2111355136 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000227_070 |
0.211704564 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 12 |
Hb_002200_050 |
0.2140682705 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_000661_080 |
0.2142466924 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 14 |
Hb_006153_030 |
0.215031566 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 15 |
Hb_001387_020 |
0.2174452217 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 16 |
Hb_005694_020 |
0.2184384569 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 17 |
Hb_010815_020 |
0.2189786318 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000160_160 |
0.2215994378 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011794_080 |
0.2217322181 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 20 |
Hb_002126_010 |
0.2218312967 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |