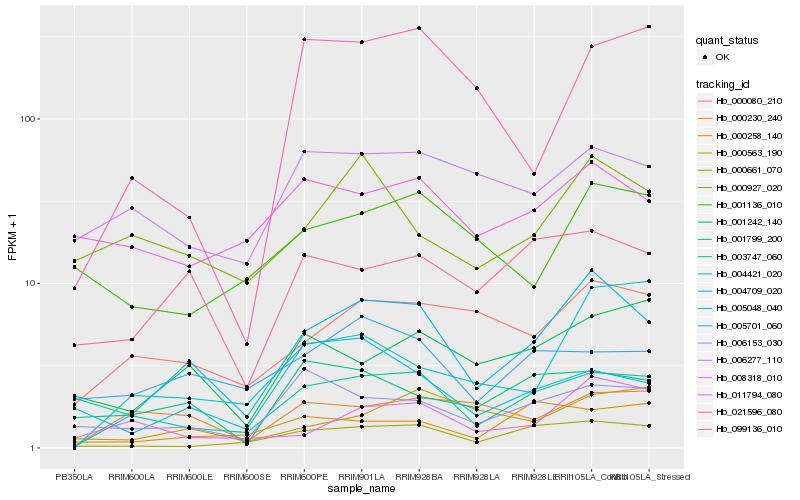
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005048_040 |
0.0 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 2 |
Hb_000661_070 |
0.194448236 |
- |
- |
- |
| 3 |
Hb_000080_210 |
0.2148258921 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 4 |
Hb_099136_010 |
0.216595882 |
- |
- |
PREDICTED: cysteine synthase-like isoform X3 [Cucumis sativus] |
| 5 |
Hb_008318_010 |
0.2396113549 |
- |
- |
PREDICTED: WD repeat-containing protein 82-B isoform X1 [Jatropha curcas] |
| 6 |
Hb_001242_140 |
0.2402532139 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 7 |
Hb_000927_020 |
0.2409723796 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 8 |
Hb_005701_060 |
0.2419330763 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 9 |
Hb_004421_020 |
0.2425849429 |
- |
- |
- |
| 10 |
Hb_000563_190 |
0.2438525417 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |
| 11 |
Hb_006153_030 |
0.2449954575 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 12 |
Hb_000230_240 |
0.2494199049 |
- |
- |
tRNA/rRNA methyltransferase family protein [Populus trichocarpa] |
| 13 |
Hb_004709_020 |
0.2495337444 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 14 |
Hb_001136_010 |
0.2503371402 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 15 |
Hb_006277_110 |
0.2509334012 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000258_140 |
0.2513498921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003747_060 |
0.2519595302 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 18 |
Hb_021596_080 |
0.2524659348 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 19 |
Hb_001799_200 |
0.2527513279 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 20 |
Hb_011794_080 |
0.2540041791 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |