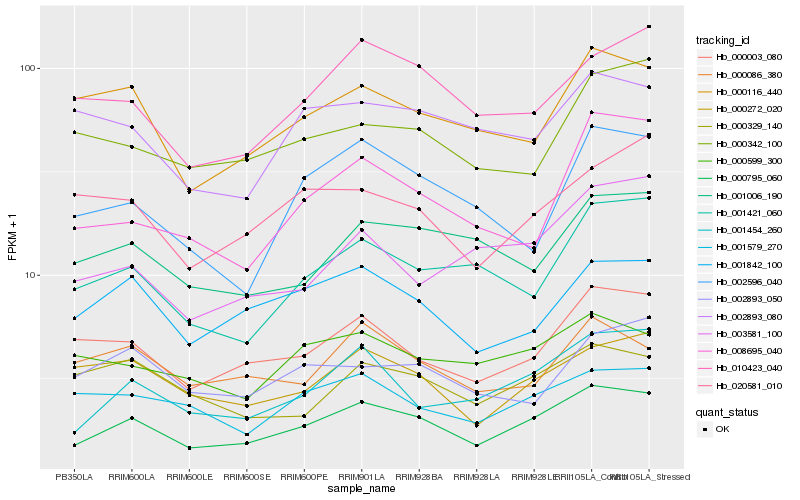
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000795_060 |
0.0 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 2 |
Hb_001454_260 |
0.0935255554 |
- |
- |
hypothetical protein RCOM_0681280 [Ricinus communis] |
| 3 |
Hb_000003_080 |
0.0991133615 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001842_100 |
0.1043062657 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |
| 5 |
Hb_010423_040 |
0.1089591905 |
- |
- |
hypothetical protein JCGZ_17463 [Jatropha curcas] |
| 6 |
Hb_000272_020 |
0.1125660044 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 7 |
Hb_001421_060 |
0.113574976 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2B isoform X2 [Jatropha curcas] |
| 8 |
Hb_001006_190 |
0.1153545737 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 9 |
Hb_000116_440 |
0.1156097645 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 6-2 [Jatropha curcas] |
| 10 |
Hb_000599_300 |
0.1159297849 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 11 |
Hb_001579_270 |
0.1167623544 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 12 |
Hb_020581_010 |
0.1213160679 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_008695_040 |
0.1220141083 |
- |
- |
PREDICTED: small ribosomal subunit protein S13, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000086_380 |
0.1234730507 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000329_140 |
0.1255702656 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000342_100 |
0.12583585 |
- |
- |
PREDICTED: PITH domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_003581_100 |
0.1260858057 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002596_040 |
0.1266366237 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002893_080 |
0.1268190557 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 20 |
Hb_002893_050 |
0.1271597529 |
- |
- |
hypothetical protein POPTR_0006s26510g [Populus trichocarpa] |