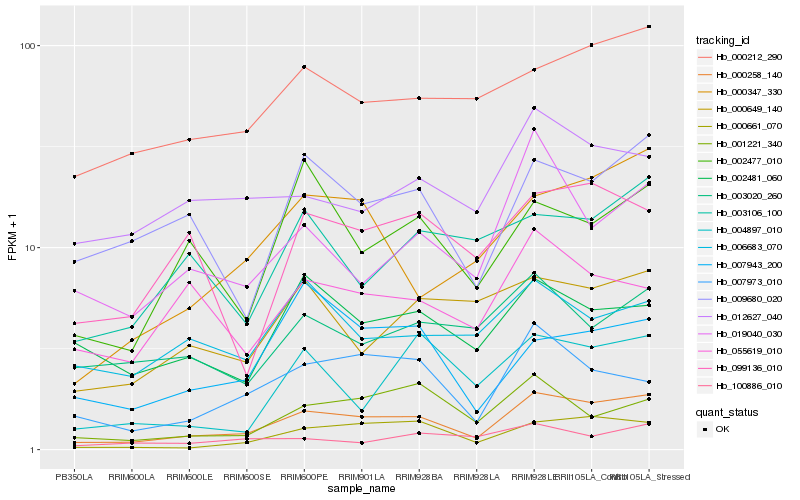
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000661_070 |
0.1513696645 |
- |
- |
- |
| 3 |
Hb_000347_330 |
0.158272477 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 4 |
Hb_000212_290 |
0.1585244608 |
- |
- |
PREDICTED: protein TIC 20-II, chloroplastic [Jatropha curcas] |
| 5 |
Hb_009680_020 |
0.1592729563 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 6 |
Hb_007973_010 |
0.163907193 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 7 |
Hb_019040_030 |
0.1712437959 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000649_140 |
0.1731973181 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002477_010 |
0.1751313337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002481_060 |
0.1776892846 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 11 |
Hb_012627_040 |
0.1779639086 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 12 |
Hb_001221_340 |
0.1787225005 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 13 |
Hb_099136_010 |
0.1791456227 |
- |
- |
PREDICTED: cysteine synthase-like isoform X3 [Cucumis sativus] |
| 14 |
Hb_003020_260 |
0.1793173715 |
- |
- |
PREDICTED: metal tolerance protein B [Jatropha curcas] |
| 15 |
Hb_003106_100 |
0.1812570083 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 16 |
Hb_055619_010 |
0.1818427561 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 17 |
Hb_007943_200 |
0.1845714323 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 18 |
Hb_006683_070 |
0.1852496422 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 19 |
Hb_004897_010 |
0.1854238707 |
- |
- |
- |
| 20 |
Hb_100886_010 |
0.1869313682 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |