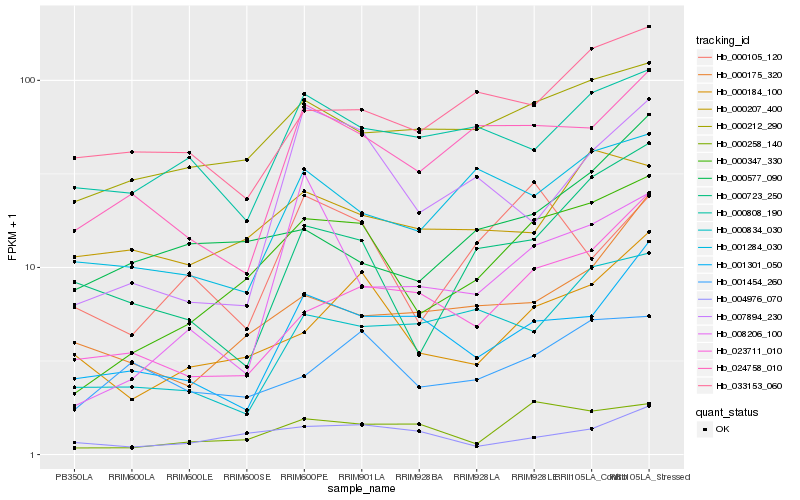
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_330 |
0.0 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 2 |
Hb_000258_140 |
0.158272477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000212_290 |
0.1588352651 |
- |
- |
PREDICTED: protein TIC 20-II, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000184_100 |
0.1655601826 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |
| 5 |
Hb_024758_010 |
0.1821023249 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 6 |
Hb_001284_030 |
0.1868593472 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_023711_010 |
0.1930319656 |
- |
- |
- |
| 8 |
Hb_001454_260 |
0.1946200252 |
- |
- |
hypothetical protein RCOM_0681280 [Ricinus communis] |
| 9 |
Hb_033153_060 |
0.1971787826 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 10 |
Hb_004976_070 |
0.198086189 |
- |
- |
eukaryotic translation initiation factor 2b, epsilon subunit, putative [Ricinus communis] |
| 11 |
Hb_000207_400 |
0.1996666635 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000723_250 |
0.2002884705 |
- |
- |
hypothetical protein JCGZ_16520 [Jatropha curcas] |
| 13 |
Hb_000834_030 |
0.2007847322 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000175_320 |
0.2008489307 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 15 |
Hb_007894_230 |
0.201763553 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 16 |
Hb_000808_190 |
0.201876714 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Eucalyptus grandis] |
| 17 |
Hb_001301_050 |
0.2028968682 |
- |
- |
- |
| 18 |
Hb_000105_120 |
0.2029641865 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 19 |
Hb_000577_090 |
0.2031638283 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_008206_100 |
0.2032939386 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |