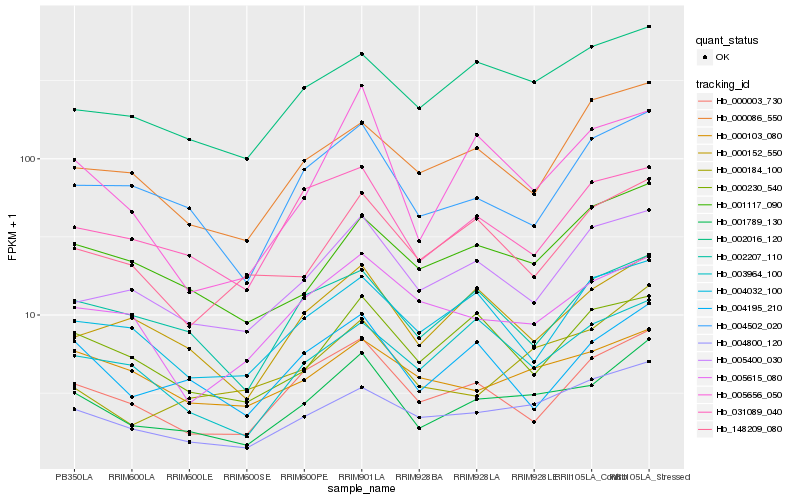
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_130 |
0.0 |
- |
- |
short-chain dehydrogenase Tic32 family protein [Populus trichocarpa] |
| 2 |
Hb_004800_120 |
0.1111388705 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000003_730 |
0.1279920397 |
- |
- |
DNA repair/transcription protein met18/mms19, putative [Ricinus communis] |
| 4 |
Hb_004502_020 |
0.1329900571 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 5 |
Hb_000152_550 |
0.133608727 |
- |
- |
PREDICTED: UMP-CMP kinase [Vitis vinifera] |
| 6 |
Hb_002016_120 |
0.1336682634 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940 [Jatropha curcas] |
| 7 |
Hb_001117_090 |
0.1343035853 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 8 |
Hb_005400_030 |
0.1366572587 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 9 |
Hb_004195_210 |
0.1368014112 |
- |
- |
- |
| 10 |
Hb_031089_040 |
0.1439159928 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 11 |
Hb_000184_100 |
0.1440362959 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |
| 12 |
Hb_148209_080 |
0.1464533952 |
- |
- |
MED32, putative [Theobroma cacao] |
| 13 |
Hb_002207_110 |
0.1466009882 |
- |
- |
- |
| 14 |
Hb_005656_050 |
0.1475324949 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 15 |
Hb_003964_100 |
0.1480432923 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 16 |
Hb_000230_540 |
0.1491259468 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 3, mitochondrial [Sesamum indicum] |
| 17 |
Hb_000086_550 |
0.1517518988 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 18 |
Hb_000103_080 |
0.1522117413 |
- |
- |
PREDICTED: ATP-dependent RNA helicase SUV3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_005615_080 |
0.1531952737 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |
| 20 |
Hb_004032_100 |
0.1539335778 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF3-like [Jatropha curcas] |