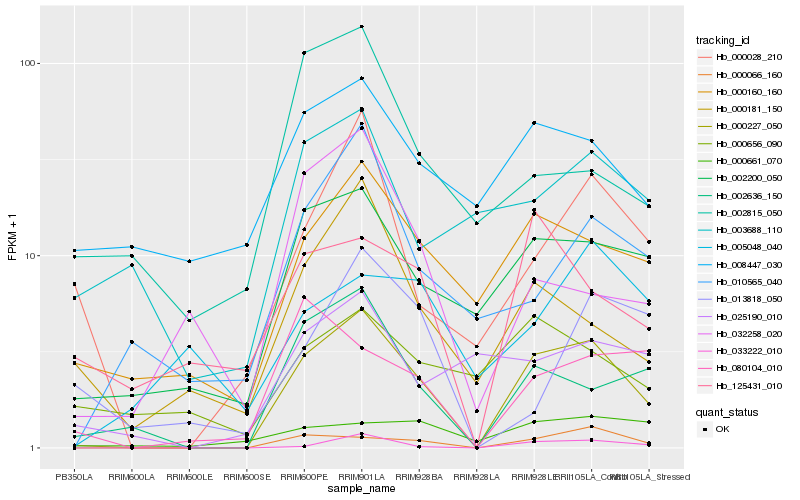
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_033222_010 |
0.2136457421 |
- |
- |
PREDICTED: uncharacterized protein LOC105173350 [Sesamum indicum] |
| 3 |
Hb_000066_160 |
0.2351212837 |
- |
- |
- |
| 4 |
Hb_002636_150 |
0.240584241 |
- |
- |
- |
| 5 |
Hb_002200_050 |
0.2520648248 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_000160_160 |
0.2549777877 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000028_210 |
0.2769608142 |
- |
- |
hypothetical protein CISIN_1g0103302mg, partial [Citrus sinensis] |
| 8 |
Hb_010565_040 |
0.2809896765 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 9 |
Hb_000181_150 |
0.2890050564 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 10 |
Hb_080104_010 |
0.3017935684 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_032258_020 |
0.3051701774 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Malus domestica] |
| 12 |
Hb_125431_010 |
0.3057953459 |
- |
- |
- |
| 13 |
Hb_025190_010 |
0.3072564208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008447_030 |
0.3120499666 |
- |
- |
- |
| 15 |
Hb_003688_110 |
0.3123612842 |
- |
- |
- |
| 16 |
Hb_000661_070 |
0.3163331072 |
- |
- |
- |
| 17 |
Hb_000656_090 |
0.3217423592 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 18 |
Hb_013818_050 |
0.3271984793 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005048_040 |
0.3272239112 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 20 |
Hb_002815_050 |
0.3274343383 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |