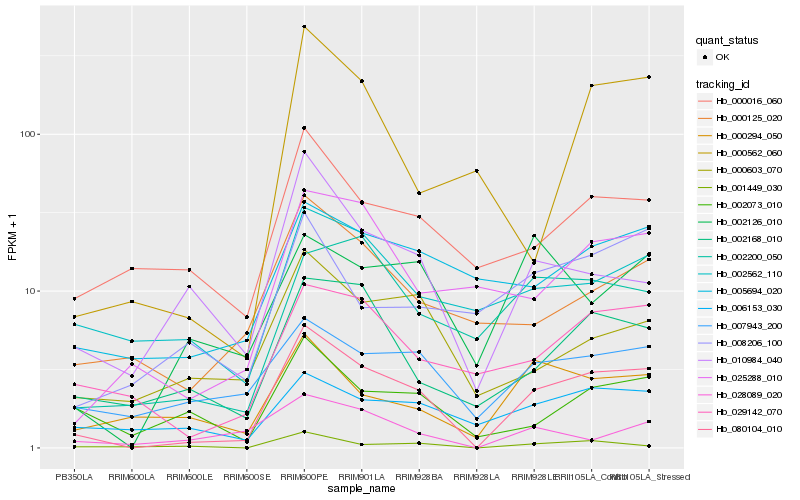
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080104_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_002168_010 |
0.2306038623 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 3 |
Hb_029142_070 |
0.2306389447 |
- |
- |
- |
| 4 |
Hb_010984_040 |
0.2371996417 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 5 |
Hb_025288_010 |
0.2408507041 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 6 |
Hb_000125_020 |
0.2419561401 |
transcription factor |
TF Family: mTERF |
- |
| 7 |
Hb_007943_200 |
0.2445042106 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_000603_070 |
0.2445329988 |
- |
- |
- |
| 9 |
Hb_002200_050 |
0.246592109 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_002073_010 |
0.2467713113 |
- |
- |
- |
| 11 |
Hb_000016_060 |
0.2468768998 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_000294_050 |
0.2484489531 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 13 |
Hb_005694_020 |
0.2494136222 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 14 |
Hb_001449_030 |
0.2497785126 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 15 |
Hb_006153_030 |
0.2557272677 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 16 |
Hb_002562_110 |
0.2565806756 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002126_010 |
0.2589960488 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 18 |
Hb_000562_060 |
0.2599851957 |
- |
- |
PREDICTED: uncharacterized protein LOC105169741 [Sesamum indicum] |
| 19 |
Hb_028089_020 |
0.2633083956 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 20 |
Hb_008206_100 |
0.2633464358 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |