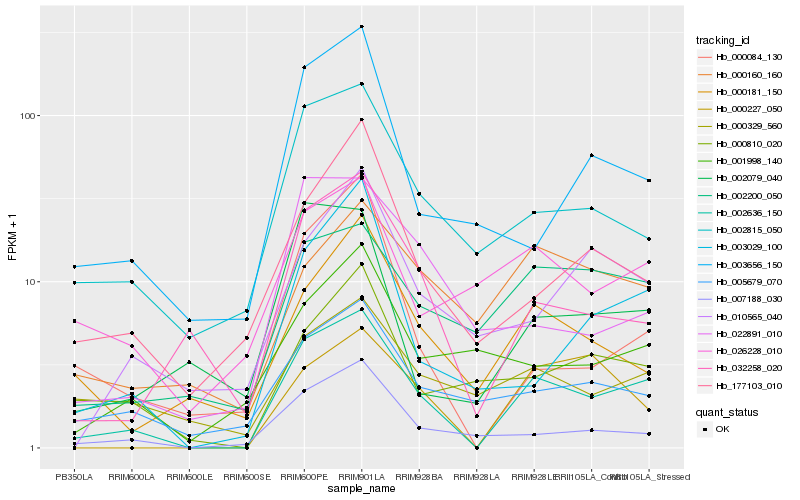
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002636_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_002815_050 |
0.2162712433 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 3 |
Hb_032258_020 |
0.2177464386 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Malus domestica] |
| 4 |
Hb_007188_030 |
0.2228910092 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 5 |
Hb_002200_050 |
0.2316259578 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_003029_100 |
0.2317499793 |
- |
- |
- |
| 7 |
Hb_005679_070 |
0.234292275 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 8 |
Hb_000181_150 |
0.2379165548 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 9 |
Hb_177103_010 |
0.2383678114 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 10 |
Hb_000810_020 |
0.2394354093 |
- |
- |
- |
| 11 |
Hb_000227_050 |
0.240584241 |
- |
- |
- |
| 12 |
Hb_000329_560 |
0.2432929179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_026228_010 |
0.2436388062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000084_130 |
0.246803915 |
- |
- |
- |
| 15 |
Hb_002079_040 |
0.2475444651 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |
| 16 |
Hb_010565_040 |
0.2487114556 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 17 |
Hb_003656_150 |
0.249375895 |
- |
- |
- |
| 18 |
Hb_000160_160 |
0.2531296821 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001998_140 |
0.253449332 |
- |
- |
- |
| 20 |
Hb_022891_010 |
0.2566584463 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |