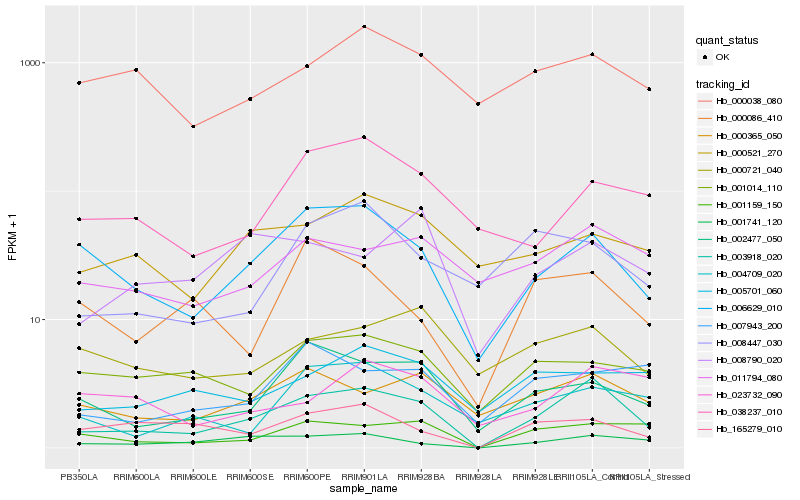
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003918_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_006629_010 |
0.1999904025 |
- |
- |
Glutathione S-transferase 2 isoform 3 [Theobroma cacao] |
| 3 |
Hb_001741_120 |
0.2103882612 |
- |
- |
hypothetical protein CARUB_v10011543mg, partial [Capsella rubella] |
| 4 |
Hb_001159_150 |
0.2165525117 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002477_050 |
0.225145023 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000365_050 |
0.2273142072 |
- |
- |
- |
| 7 |
Hb_011794_080 |
0.2372434849 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 8 |
Hb_165279_010 |
0.2456646432 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 9 |
Hb_004709_020 |
0.2460550582 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 10 |
Hb_001014_110 |
0.2500133993 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 11 |
Hb_038237_010 |
0.2505281649 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 12 |
Hb_000721_040 |
0.251090082 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005701_060 |
0.2511777748 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 14 |
Hb_007943_200 |
0.2516710577 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_000038_080 |
0.2525743009 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 16 |
Hb_000521_270 |
0.2529035438 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 17 |
Hb_008447_030 |
0.2530828925 |
- |
- |
- |
| 18 |
Hb_023732_090 |
0.2540354107 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_008790_020 |
0.2548144806 |
- |
- |
PREDICTED: uncharacterized protein LOC105638122 [Jatropha curcas] |
| 20 |
Hb_000086_410 |
0.2561921992 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |