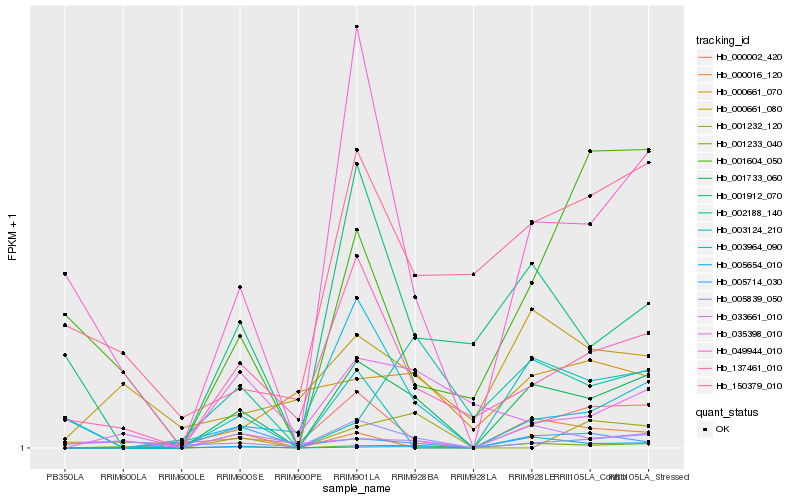
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001733_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104431099 [Eucalyptus grandis] |
| 2 |
Hb_001233_040 |
0.2457730061 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 3 |
Hb_000002_420 |
0.2600008494 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1-like [Citrus sinensis] |
| 4 |
Hb_049944_010 |
0.2754388096 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 5 |
Hb_137461_010 |
0.2950619697 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 6 |
Hb_001912_070 |
0.3075596832 |
- |
- |
hypothetical protein POPTR_0003s03380g [Populus trichocarpa] |
| 7 |
Hb_000016_120 |
0.3209004856 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 8 |
Hb_002188_140 |
0.3300774398 |
- |
- |
PREDICTED: uncharacterized protein LOC105650814 [Jatropha curcas] |
| 9 |
Hb_003964_090 |
0.3306272694 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 10 |
Hb_003124_210 |
0.3356807348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005839_050 |
0.3404290357 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_000661_070 |
0.3451460485 |
- |
- |
- |
| 13 |
Hb_005714_030 |
0.3478148631 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 14 |
Hb_033661_010 |
0.3560145442 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 15 |
Hb_035398_010 |
0.3560827203 |
- |
- |
hypothetical protein JCGZ_13029 [Jatropha curcas] |
| 16 |
Hb_001232_120 |
0.3596684293 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 17 |
Hb_005654_010 |
0.3601245317 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_001604_050 |
0.3612133249 |
- |
- |
PREDICTED: uncharacterized protein LOC105639208 [Jatropha curcas] |
| 19 |
Hb_000661_080 |
0.3627020126 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 20 |
Hb_150379_010 |
0.3645727997 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |