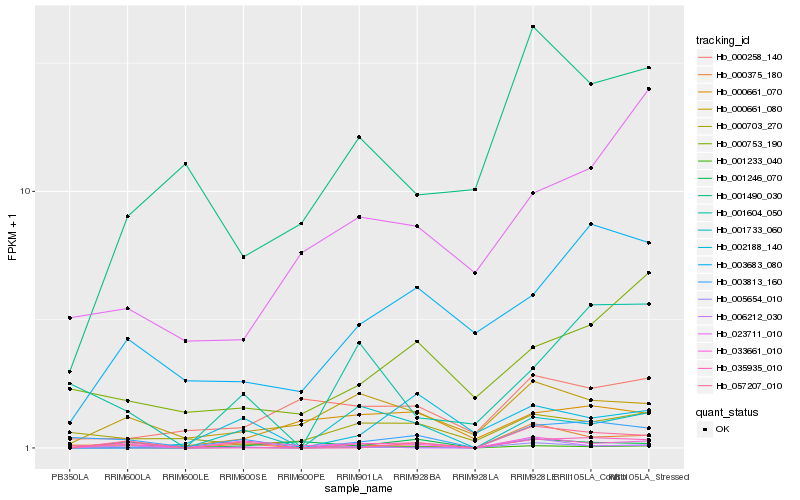
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001233_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 2 |
Hb_035935_010 |
0.2178331995 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 3 |
Hb_001733_060 |
0.2457730061 |
- |
- |
PREDICTED: uncharacterized protein LOC104431099 [Eucalyptus grandis] |
| 4 |
Hb_003813_160 |
0.2705337712 |
- |
- |
- |
| 5 |
Hb_033661_010 |
0.2797432374 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 6 |
Hb_000661_080 |
0.2865372942 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 7 |
Hb_057207_010 |
0.2904983457 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 8 |
Hb_005654_010 |
0.2937316828 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_003683_080 |
0.3193318737 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 10 |
Hb_000703_270 |
0.3274390306 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 11 |
Hb_001490_030 |
0.3311027588 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000753_190 |
0.33274434 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 13 |
Hb_002188_140 |
0.3345110882 |
- |
- |
PREDICTED: uncharacterized protein LOC105650814 [Jatropha curcas] |
| 14 |
Hb_000375_180 |
0.33563896 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 15 |
Hb_001604_050 |
0.3385444163 |
- |
- |
PREDICTED: uncharacterized protein LOC105639208 [Jatropha curcas] |
| 16 |
Hb_006212_030 |
0.3420742679 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 17 |
Hb_001246_070 |
0.3437187103 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000661_070 |
0.3439956273 |
- |
- |
- |
| 19 |
Hb_000258_140 |
0.34440589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_023711_010 |
0.3461574785 |
- |
- |
- |