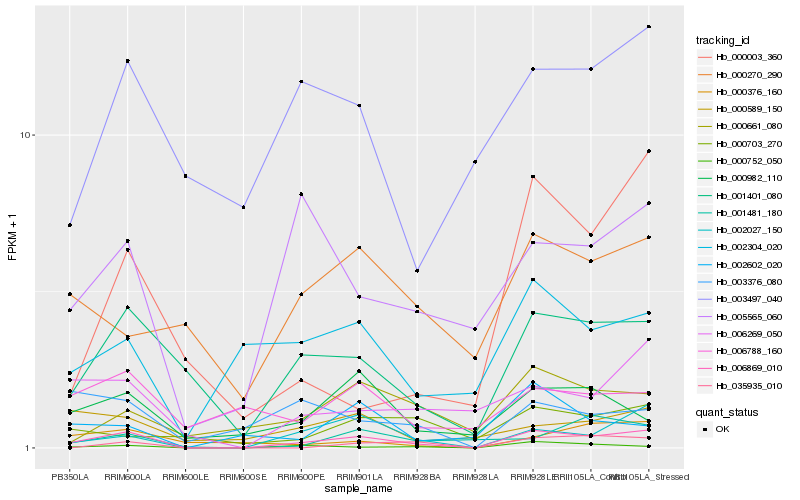
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006869_010 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_002027_150 |
0.2314449592 |
- |
- |
- |
| 3 |
Hb_001401_080 |
0.2400324765 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000752_050 |
0.2675138378 |
- |
- |
PREDICTED: uncharacterized protein LOC102610887 [Citrus sinensis] |
| 5 |
Hb_006269_050 |
0.2800600085 |
- |
- |
hypothetical protein POPTR_0004s13450g [Populus trichocarpa] |
| 6 |
Hb_005565_060 |
0.2809301762 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000982_110 |
0.2874804637 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_002304_020 |
0.2938323889 |
- |
- |
- |
| 9 |
Hb_035935_010 |
0.3027997541 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 10 |
Hb_000661_080 |
0.3030039719 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 11 |
Hb_000589_150 |
0.3040688182 |
- |
- |
PREDICTED: factor of DNA methylation 1-like isoform X1 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_000376_160 |
0.3050096213 |
- |
- |
- |
| 13 |
Hb_003376_080 |
0.3080371708 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002602_020 |
0.3088362753 |
- |
- |
- |
| 15 |
Hb_006788_160 |
0.3127300959 |
- |
- |
hypothetical protein POPTR_0002s04680g [Populus trichocarpa] |
| 16 |
Hb_001481_180 |
0.3140422469 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 17 |
Hb_000003_360 |
0.3149007683 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 18 |
Hb_003497_040 |
0.3151743803 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000270_290 |
0.315333118 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 20 |
Hb_000703_270 |
0.3157857319 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |