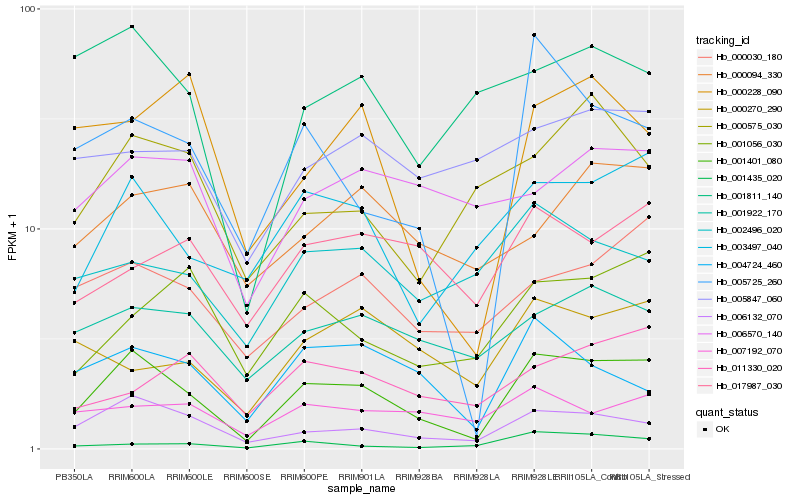
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001401_080 |
0.0 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006132_070 |
0.1560151898 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_003497_040 |
0.1783687293 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_005725_260 |
0.1934930764 |
- |
- |
squalene monooxygenase [Jatropha curcas] |
| 5 |
Hb_000030_180 |
0.1955822437 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001922_170 |
0.2025606765 |
- |
- |
Plastidic pyruvate kinase beta subunit 1 isoform 2 [Theobroma cacao] |
| 7 |
Hb_004724_460 |
0.2059283431 |
- |
- |
hypothetical protein EUGRSUZ_L02100 [Eucalyptus grandis] |
| 8 |
Hb_001056_030 |
0.2086077919 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 9 |
Hb_007192_070 |
0.2132632179 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_017987_030 |
0.2157679831 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 11 |
Hb_001811_140 |
0.2163061332 |
- |
- |
hypothetical protein POPTR_0007s03140g [Populus trichocarpa] |
| 12 |
Hb_002496_020 |
0.2166877958 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000228_090 |
0.217582266 |
- |
- |
PREDICTED: protein PAM68, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000094_330 |
0.2184861896 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 15 |
Hb_000270_290 |
0.2208484212 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 16 |
Hb_011330_020 |
0.2214557858 |
transcription factor |
TF Family: ARID |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000575_030 |
0.2216472316 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001435_020 |
0.221699076 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 19 |
Hb_006570_140 |
0.2222117104 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 20 |
Hb_005847_060 |
0.2225806232 |
- |
- |
hypothetical protein POPTR_0006s02920g [Populus trichocarpa] |