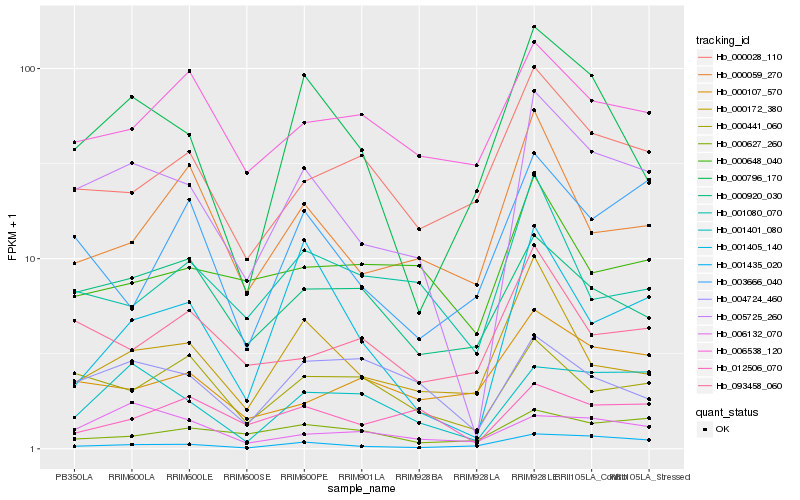
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_260 |
0.0 |
- |
- |
squalene monooxygenase [Jatropha curcas] |
| 2 |
Hb_001401_080 |
0.1934930764 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000441_060 |
0.1996060438 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 4 |
Hb_000172_380 |
0.2000716136 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 5 |
Hb_000796_170 |
0.2059615761 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 6 |
Hb_000028_110 |
0.2088468867 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 7 |
Hb_001435_020 |
0.2088921813 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 8 |
Hb_012506_070 |
0.2090080748 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 9 |
Hb_000920_030 |
0.2093595917 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004724_460 |
0.2118623777 |
- |
- |
hypothetical protein EUGRSUZ_L02100 [Eucalyptus grandis] |
| 11 |
Hb_000059_270 |
0.2120529743 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 12 |
Hb_093458_060 |
0.2122328738 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 13 |
Hb_001405_140 |
0.2142808613 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 14 |
Hb_001080_070 |
0.2160635558 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 15 |
Hb_006538_120 |
0.2181390364 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000627_260 |
0.2208436881 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 17 |
Hb_000648_040 |
0.2216044127 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_000107_570 |
0.2226475159 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003666_040 |
0.2231982547 |
- |
- |
PREDICTED: uncharacterized protein LOC105639624 [Jatropha curcas] |
| 20 |
Hb_006132_070 |
0.2242176788 |
- |
- |
phosphate transporter [Manihot esculenta] |