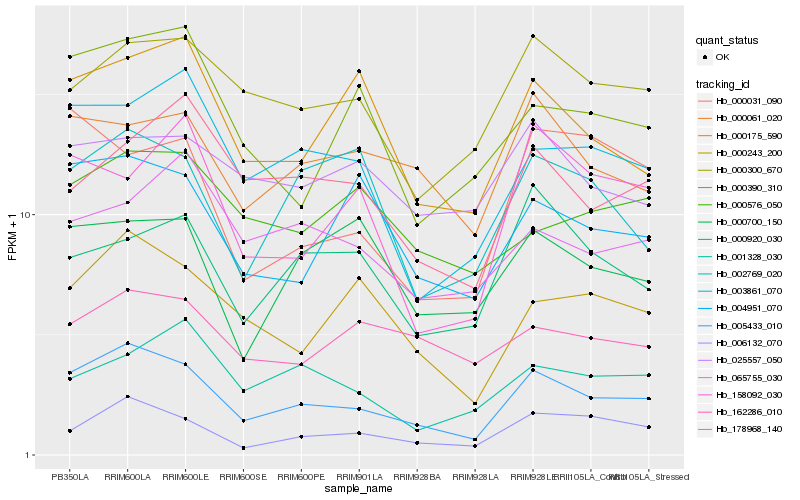
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005433_010 |
0.0 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 2 |
Hb_006132_070 |
0.1247919354 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_000243_200 |
0.1330107182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003861_070 |
0.136159506 |
- |
- |
PREDICTED: uncharacterized protein LOC105650781 [Jatropha curcas] |
| 5 |
Hb_004951_070 |
0.1400160285 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 6 |
Hb_000175_590 |
0.1440529915 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000300_670 |
0.1453027779 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000061_020 |
0.1475617939 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 9 |
Hb_000390_310 |
0.1501108229 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002769_020 |
0.1559260165 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 11 |
Hb_178968_140 |
0.1571261172 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000031_090 |
0.1577401478 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g10690 [Jatropha curcas] |
| 13 |
Hb_158092_030 |
0.1618210088 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP19, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000700_150 |
0.162285438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000920_030 |
0.1624873264 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001328_030 |
0.1629205555 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 17 |
Hb_000576_050 |
0.16353354 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 18 |
Hb_162286_010 |
0.1654871585 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 19 |
Hb_065755_030 |
0.1663798834 |
- |
- |
- |
| 20 |
Hb_025557_050 |
0.1667731804 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |