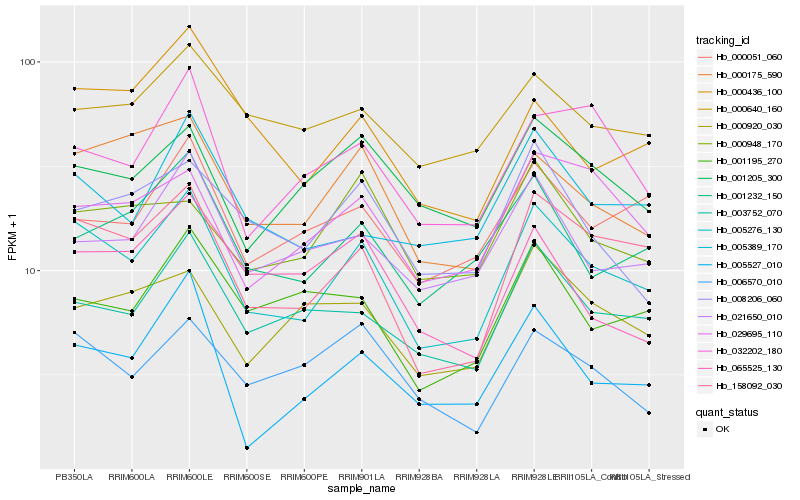
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005276_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_158092_030 |
0.0805423549 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP19, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000175_590 |
0.1139062445 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003752_070 |
0.1169141465 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001232_150 |
0.1227472153 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 6 |
Hb_021650_010 |
0.1262415464 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 7 |
Hb_005527_010 |
0.1268561305 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_005389_170 |
0.1270014301 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 9 |
Hb_000051_060 |
0.1277937238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_029695_110 |
0.1292928558 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 11 |
Hb_006570_010 |
0.1300734608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_032202_180 |
0.1319020198 |
- |
- |
PREDICTED: protein-ribulosamine 3-kinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001205_300 |
0.1320110678 |
- |
- |
glutamyl-tRNA synthetase 1, 2, putative [Ricinus communis] |
| 14 |
Hb_008206_060 |
0.135385499 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000436_100 |
0.1356550111 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 16 |
Hb_000920_030 |
0.1358007303 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001195_270 |
0.1361753875 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000640_160 |
0.1364319523 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 19 |
Hb_000948_170 |
0.1404121511 |
- |
- |
hypothetical protein JCGZ_06749 [Jatropha curcas] |
| 20 |
Hb_065525_130 |
0.1407179187 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |