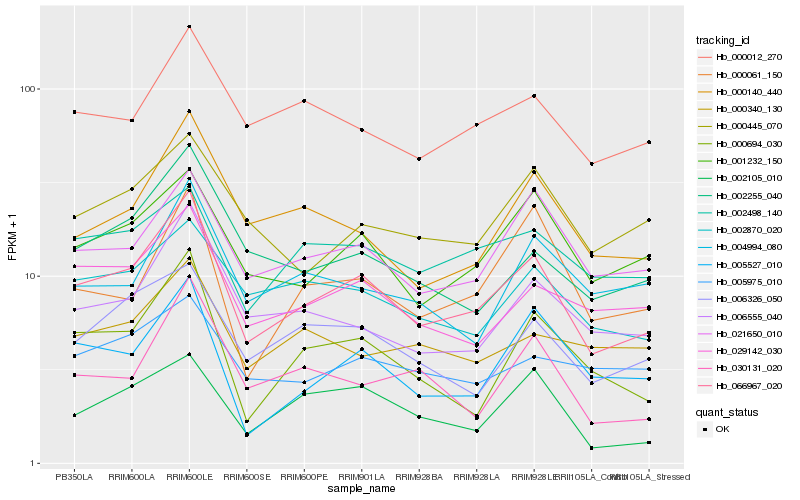
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066967_020 |
0.0 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |
| 2 |
Hb_021650_010 |
0.1159064899 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 3 |
Hb_006555_040 |
0.1161986577 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 4 |
Hb_001232_150 |
0.1166051749 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 5 |
Hb_029142_030 |
0.1166681115 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 6 |
Hb_000140_440 |
0.1209574614 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 7 |
Hb_000694_030 |
0.1227331327 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 8 |
Hb_002255_040 |
0.1234371576 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 9 |
Hb_000012_270 |
0.1248787659 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 10 |
Hb_006326_050 |
0.1258487912 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_002870_020 |
0.1273636737 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005527_010 |
0.1276040505 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002498_140 |
0.1328080562 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 14 |
Hb_000061_150 |
0.1348270624 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002105_010 |
0.1367798955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004994_080 |
0.1376958755 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000445_070 |
0.1377911189 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 18 |
Hb_005975_010 |
0.1399774338 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_030131_020 |
0.1407632302 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 20 |
Hb_000340_130 |
0.1431386184 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |