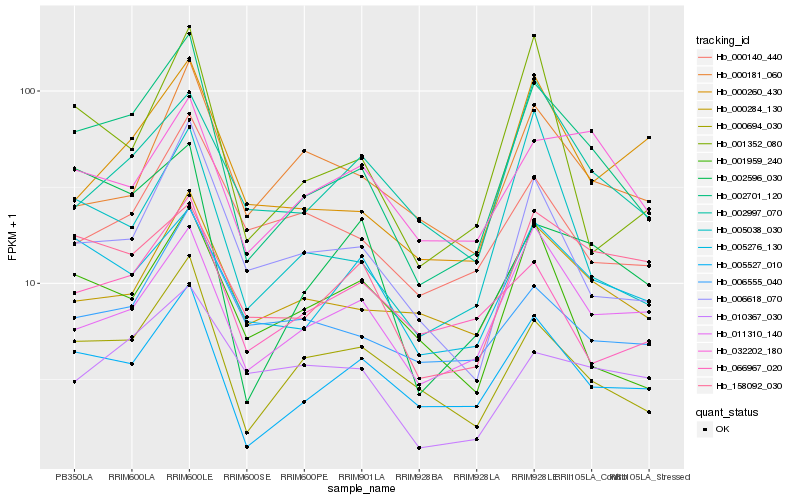
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 2 |
Hb_005527_010 |
0.1268283076 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000694_030 |
0.1284386923 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 4 |
Hb_006618_070 |
0.1453799727 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011310_140 |
0.165622597 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 6 |
Hb_000284_130 |
0.1656908359 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 7 |
Hb_005038_030 |
0.1673370203 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005276_130 |
0.1679179944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001352_080 |
0.1698266136 |
- |
- |
PREDICTED: uncharacterized protein At5g02240 [Jatropha curcas] |
| 10 |
Hb_002596_030 |
0.1728089292 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 11 |
Hb_006555_040 |
0.1738066504 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 12 |
Hb_158092_030 |
0.1752791801 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP19, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000260_430 |
0.177978549 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 14 |
Hb_066967_020 |
0.1797198098 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |
| 15 |
Hb_001959_240 |
0.1806813127 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_000140_440 |
0.1813711762 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 17 |
Hb_032202_180 |
0.181686753 |
- |
- |
PREDICTED: protein-ribulosamine 3-kinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002997_070 |
0.1819826114 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 19 |
Hb_010367_030 |
0.1838456113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000181_060 |
0.1840226292 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |