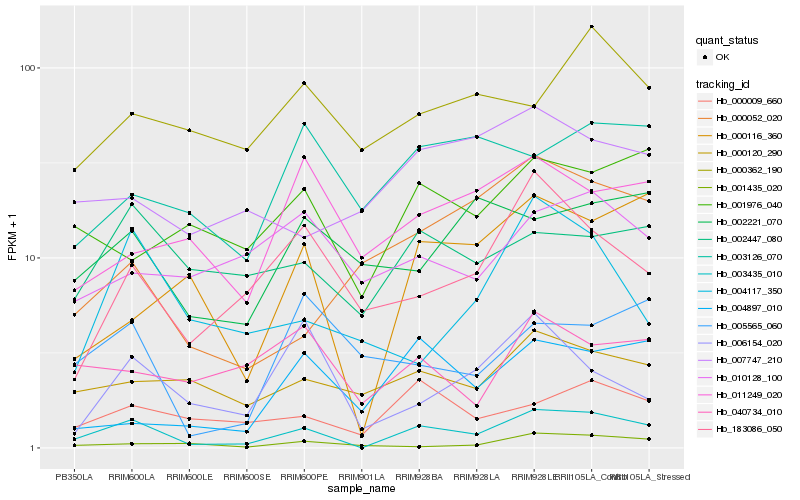
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003435_010 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABC2a-like [Jatropha curcas] |
| 2 |
Hb_000009_660 |
0.2363972019 |
- |
- |
- |
| 3 |
Hb_000116_360 |
0.2420419858 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 4 |
Hb_000120_290 |
0.248969586 |
- |
- |
PREDICTED: N-acylphosphatidylethanolamine synthase isoform X1 [Jatropha curcas] |
| 5 |
Hb_002447_080 |
0.2496742439 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 6 |
Hb_183086_050 |
0.2504001556 |
- |
- |
PREDICTED: metal-nicotianamine transporter YSL3 [Jatropha curcas] |
| 7 |
Hb_001976_040 |
0.2581970655 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_040734_010 |
0.2668188704 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 9 |
Hb_001435_020 |
0.2706243598 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 10 |
Hb_004117_350 |
0.2709439536 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 11 |
Hb_002221_070 |
0.2709711474 |
- |
- |
beta-glucanase, putative [Ricinus communis] |
| 12 |
Hb_007747_210 |
0.27283445 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_003126_070 |
0.2739337267 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010128_100 |
0.2747625041 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000362_190 |
0.2751387691 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 16 |
Hb_004897_010 |
0.2754466644 |
- |
- |
- |
| 17 |
Hb_011249_020 |
0.2756947113 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 18 |
Hb_000052_020 |
0.2766896083 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 19 |
Hb_006154_020 |
0.2771072739 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 20 |
Hb_005565_060 |
0.2773969343 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 isoform X1 [Jatropha curcas] |