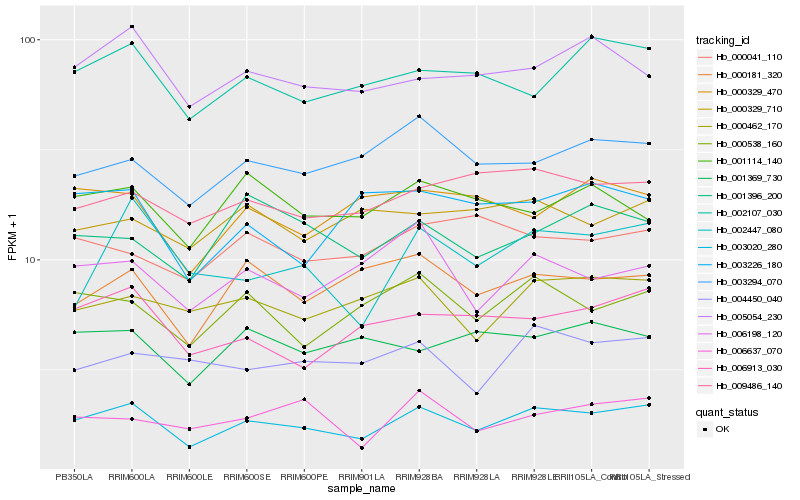
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_280 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa026521mg [Prunus persica] |
| 2 |
Hb_000538_160 |
0.0960154215 |
- |
- |
PREDICTED: RNA-binding protein 5 isoform X3 [Jatropha curcas] |
| 3 |
Hb_006913_030 |
0.0984487592 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 4 |
Hb_002107_030 |
0.0984515623 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005054_230 |
0.0989978163 |
- |
- |
PREDICTED: wiskott-Aldrich syndrome protein family member 2 [Jatropha curcas] |
| 6 |
Hb_000462_170 |
0.1010245397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003294_070 |
0.1013033664 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 8 |
Hb_009486_140 |
0.1020335156 |
- |
- |
PREDICTED: uncharacterized protein LOC105643248 [Jatropha curcas] |
| 9 |
Hb_000181_320 |
0.1045295098 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_006198_120 |
0.1073996367 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 11 |
Hb_004450_040 |
0.1094419758 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 12 |
Hb_001396_200 |
0.1101258415 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002447_080 |
0.1109572589 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 14 |
Hb_000329_470 |
0.111363509 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 15 |
Hb_001114_140 |
0.1137897372 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 16 |
Hb_001369_730 |
0.1138490696 |
- |
- |
F5O11.10, putative isoform 1 [Theobroma cacao] |
| 17 |
Hb_003226_180 |
0.1141462483 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 18 |
Hb_006637_070 |
0.1144129257 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 19 |
Hb_000329_710 |
0.1156249207 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000041_110 |
0.1164012681 |
- |
- |
PREDICTED: WW domain-binding protein 11 isoform X1 [Jatropha curcas] |