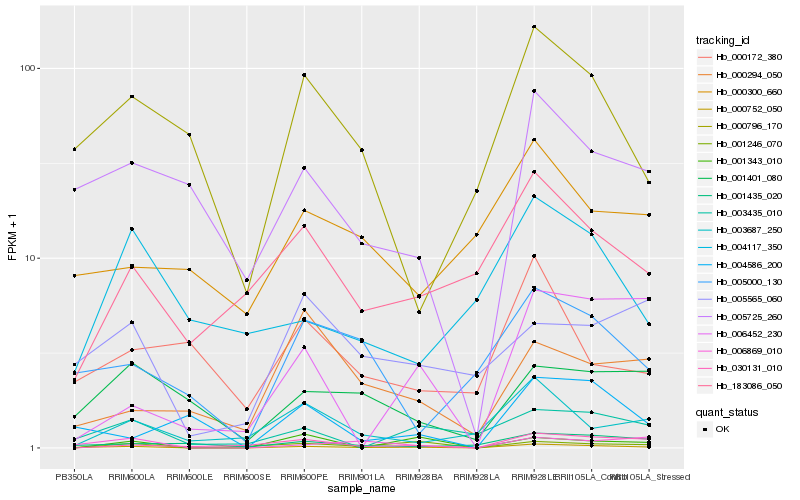
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000752_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102610887 [Citrus sinensis] |
| 2 |
Hb_005725_260 |
0.2672814416 |
- |
- |
squalene monooxygenase [Jatropha curcas] |
| 3 |
Hb_006869_010 |
0.2675138378 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_030131_010 |
0.2879056517 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 5 |
Hb_003435_010 |
0.2932528527 |
- |
- |
PREDICTED: ras-related protein RABC2a-like [Jatropha curcas] |
| 6 |
Hb_000796_170 |
0.2988504097 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 7 |
Hb_004586_200 |
0.2992754219 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 8 |
Hb_001246_070 |
0.3008298754 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001435_020 |
0.3040916681 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 10 |
Hb_001401_080 |
0.3042278986 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005000_130 |
0.3079754241 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 12 |
Hb_183086_050 |
0.3080368001 |
- |
- |
PREDICTED: metal-nicotianamine transporter YSL3 [Jatropha curcas] |
| 13 |
Hb_005565_060 |
0.3143123607 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006452_230 |
0.3198264154 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 15 |
Hb_003687_250 |
0.3217573075 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 16 |
Hb_001343_010 |
0.323369482 |
- |
- |
- |
| 17 |
Hb_000172_380 |
0.3323365532 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 18 |
Hb_004117_350 |
0.3334429065 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 19 |
Hb_000300_660 |
0.3361238622 |
- |
- |
unknown [Glycine max] |
| 20 |
Hb_000294_050 |
0.3364033288 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |