| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
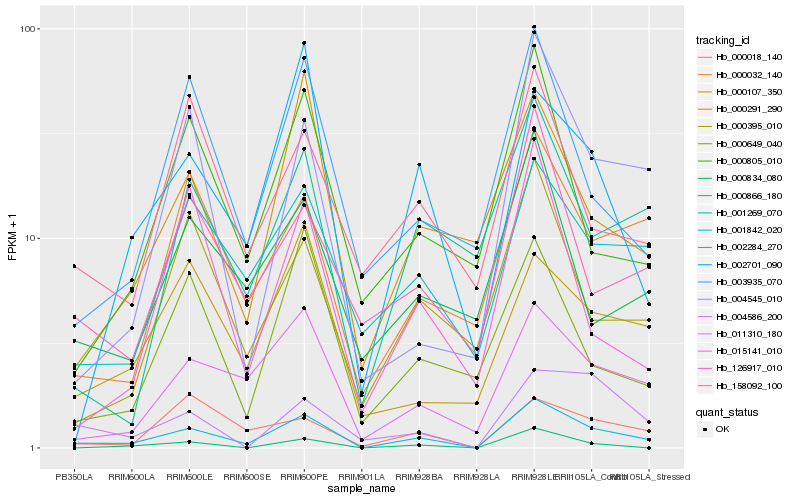
Hb_002284_270 |
0.0 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_011310_180 |
0.1895377508 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001842_020 |
0.2168634377 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 4 |
Hb_004545_010 |
0.2188953271 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 5 |
Hb_003935_070 |
0.2207640781 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000032_140 |
0.2215414207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_126917_010 |
0.2268309567 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 8 |
Hb_000107_350 |
0.228367009 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 9 |
Hb_000395_010 |
0.2296512427 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 10 |
Hb_000805_010 |
0.2301900561 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 11 |
Hb_000291_290 |
0.2326725067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002701_090 |
0.2328842536 |
- |
- |
hypothetical protein POPTR_0017s13430g [Populus trichocarpa] |
| 13 |
Hb_000018_140 |
0.2337904967 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 14 |
Hb_000866_180 |
0.2357873171 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 15 |
Hb_000649_040 |
0.235799476 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 16 |
Hb_000834_080 |
0.2368045958 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 17 |
Hb_001269_070 |
0.2368359837 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_004586_200 |
0.2378856305 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 19 |
Hb_158092_100 |
0.237923826 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 20 |
Hb_015141_010 |
0.2380191437 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |