| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
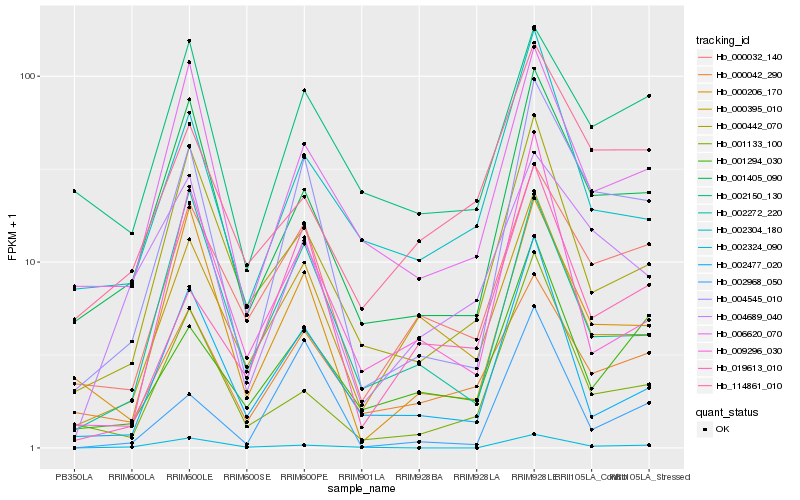
Hb_004545_010 |
0.0 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 2 |
Hb_001405_090 |
0.1521718802 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000032_140 |
0.1733916718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000206_170 |
0.175685805 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 5 |
Hb_006620_070 |
0.1873712145 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 6 |
Hb_000042_290 |
0.1929954478 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 7 |
Hb_019613_010 |
0.1982102659 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_004689_040 |
0.1995291633 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 9 |
Hb_000442_070 |
0.1998154451 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 10 |
Hb_001294_030 |
0.202813044 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 11 |
Hb_000395_010 |
0.2064116503 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 12 |
Hb_002304_180 |
0.2086858244 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002477_020 |
0.2094932396 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 14 |
Hb_001133_100 |
0.2105656556 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 15 |
Hb_002150_130 |
0.2105901973 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_002272_220 |
0.2140571918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002324_090 |
0.2151859576 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 18 |
Hb_009296_030 |
0.2160393871 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 19 |
Hb_114861_010 |
0.2161203473 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 20 |
Hb_002968_050 |
0.2172348641 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |