| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
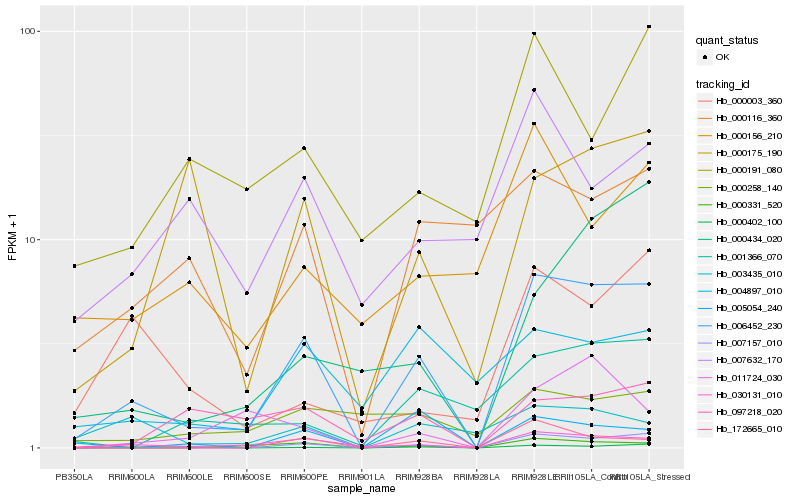
Hb_006452_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 2 |
Hb_007157_010 |
0.192326976 |
- |
- |
- |
| 3 |
Hb_030131_010 |
0.2024110929 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 4 |
Hb_000331_520 |
0.2387323049 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 5 |
Hb_000402_100 |
0.2569728993 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 6 |
Hb_172665_010 |
0.2653454331 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_001366_070 |
0.2812174625 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000434_020 |
0.2947823778 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 9 |
Hb_000116_360 |
0.2963332564 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 10 |
Hb_000175_190 |
0.2976986971 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 11 |
Hb_097218_020 |
0.2996725053 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 12 |
Hb_004897_010 |
0.3023354914 |
- |
- |
- |
| 13 |
Hb_000156_210 |
0.3026387027 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 14 |
Hb_000003_360 |
0.3040077381 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 15 |
Hb_000191_080 |
0.3056670775 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 16 |
Hb_000258_140 |
0.3065257974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011724_030 |
0.3070725302 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 18 |
Hb_005054_240 |
0.3089642862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003435_010 |
0.3095430097 |
- |
- |
PREDICTED: ras-related protein RABC2a-like [Jatropha curcas] |
| 20 |
Hb_007632_170 |
0.3117533103 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |