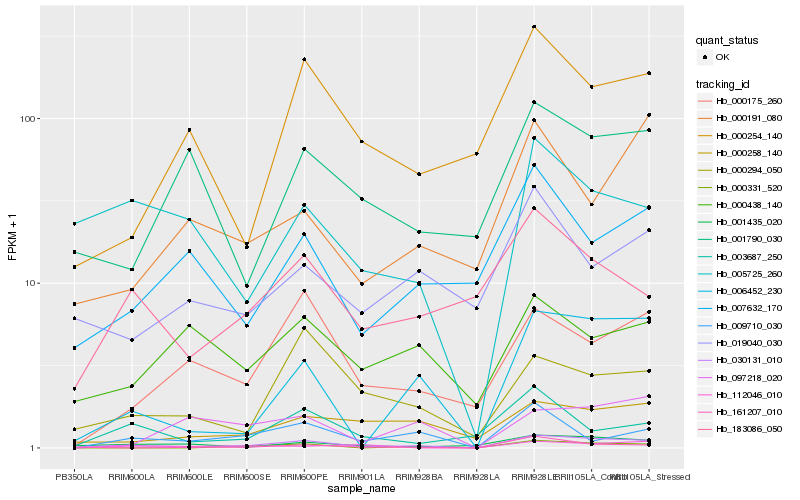
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030131_010 |
0.0 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 2 |
Hb_006452_230 |
0.2024110929 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 3 |
Hb_000331_520 |
0.23754026 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 4 |
Hb_000175_260 |
0.2380163497 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 5 |
Hb_009710_030 |
0.2533871267 |
- |
- |
- |
| 6 |
Hb_000254_140 |
0.2554236835 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000258_140 |
0.2567567774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_183086_050 |
0.2595286231 |
- |
- |
PREDICTED: metal-nicotianamine transporter YSL3 [Jatropha curcas] |
| 9 |
Hb_000294_050 |
0.2598008378 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 10 |
Hb_001435_020 |
0.2604236667 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 11 |
Hb_007632_170 |
0.2667921394 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 12 |
Hb_161207_010 |
0.2689540753 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 13 |
Hb_097218_020 |
0.2690915245 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 14 |
Hb_003687_250 |
0.2728043259 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 15 |
Hb_112046_010 |
0.2764263446 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 16 |
Hb_001790_030 |
0.2825139592 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 17 |
Hb_019040_030 |
0.2852546009 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005725_260 |
0.2856368129 |
- |
- |
squalene monooxygenase [Jatropha curcas] |
| 19 |
Hb_000438_140 |
0.2860728736 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 20 |
Hb_000191_080 |
0.2875485994 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |