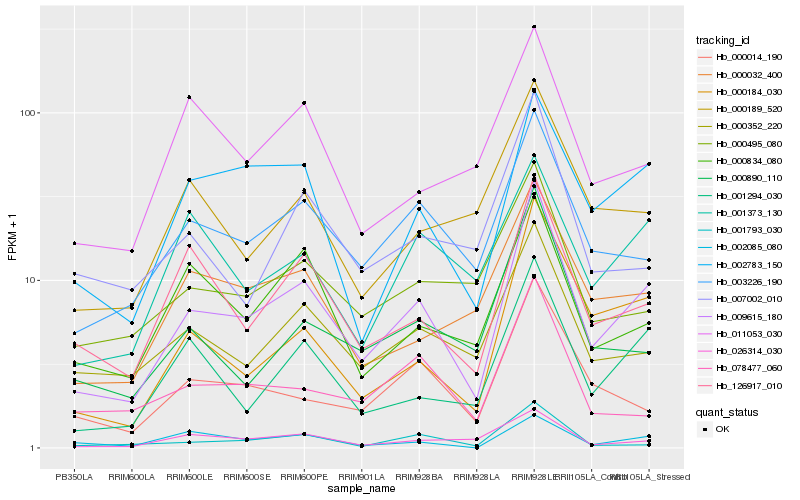
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009615_180 |
0.0 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 2 |
Hb_003226_190 |
0.1504354616 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 3 |
Hb_000184_030 |
0.1531087097 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002783_150 |
0.1569484724 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 5 |
Hb_001294_030 |
0.1584562144 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 6 |
Hb_126917_010 |
0.1592760459 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 7 |
Hb_000352_220 |
0.1641095311 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000032_400 |
0.1755306888 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 9 |
Hb_001793_030 |
0.1770873182 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 10 |
Hb_026314_030 |
0.1785886651 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 11 |
Hb_000890_110 |
0.178600775 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 12 |
Hb_000014_190 |
0.1808594472 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_000495_080 |
0.1821864467 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 14 |
Hb_000834_080 |
0.1834400657 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 15 |
Hb_000189_520 |
0.1840030747 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 16 |
Hb_078477_060 |
0.1885964745 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 17 |
Hb_002085_080 |
0.1888280443 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Jatropha curcas] |
| 18 |
Hb_007002_010 |
0.1897791933 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 19 |
Hb_011053_030 |
0.193487854 |
- |
- |
- |
| 20 |
Hb_001373_130 |
0.1938456601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |