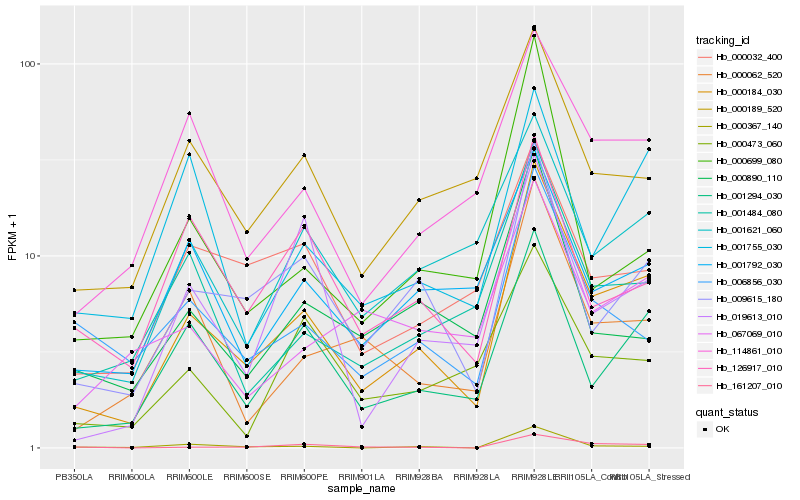
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000184_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 2 |
Hb_009615_180 |
0.1531087097 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 3 |
Hb_001294_030 |
0.1563216632 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 4 |
Hb_000062_520 |
0.1794104152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000189_520 |
0.1806424395 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 6 |
Hb_001484_080 |
0.1817317324 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 7 |
Hb_114861_010 |
0.1820493199 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 8 |
Hb_161207_010 |
0.1835791379 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 9 |
Hb_001621_060 |
0.184106042 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006856_030 |
0.1915105934 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 11 |
Hb_001792_030 |
0.1940515206 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 12 |
Hb_000890_110 |
0.1972274425 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 13 |
Hb_000032_400 |
0.2000949241 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 14 |
Hb_000699_080 |
0.2041344437 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000367_140 |
0.207031896 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 16 |
Hb_019613_010 |
0.2076166941 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_126917_010 |
0.2080835563 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 18 |
Hb_001755_030 |
0.2089898102 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_067069_010 |
0.2108725289 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 20 |
Hb_000473_060 |
0.2109032618 |
- |
- |
PREDICTED: psbP domain-containing protein 2, chloroplastic [Jatropha curcas] |