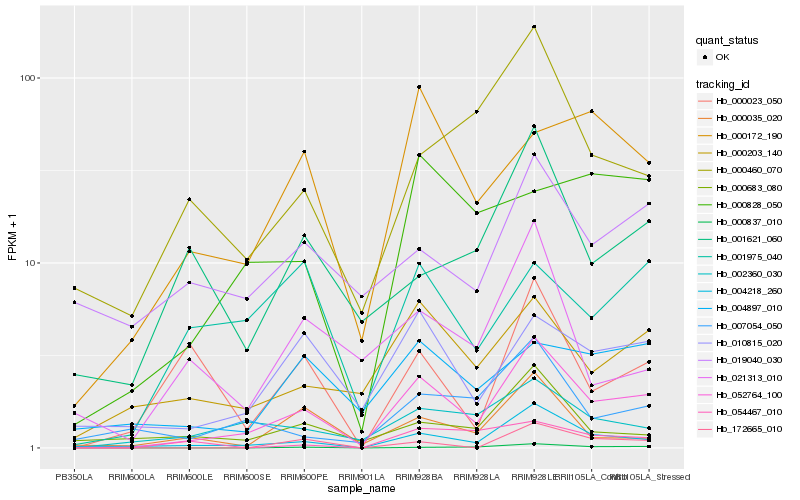
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052764_100 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase 8-like [Jatropha curcas] |
| 2 |
Hb_004218_260 |
0.1643337989 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 3 |
Hb_010815_020 |
0.2266263628 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000203_140 |
0.2337901895 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 5 |
Hb_002360_030 |
0.2339844383 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 6 |
Hb_000837_010 |
0.2431657943 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_007054_050 |
0.2495027738 |
- |
- |
- |
| 8 |
Hb_000460_070 |
0.2516042473 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 9 |
Hb_000683_080 |
0.261315343 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 10 |
Hb_000035_020 |
0.2613349473 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 11 |
Hb_000172_190 |
0.2616645366 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 12 |
Hb_172665_010 |
0.2625909825 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_001621_060 |
0.2669975345 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004897_010 |
0.2674179851 |
- |
- |
- |
| 15 |
Hb_000828_050 |
0.2716841621 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_021313_010 |
0.2719187878 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 17 |
Hb_000023_050 |
0.2752336133 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 18 |
Hb_001975_040 |
0.2755567061 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 19 |
Hb_019040_030 |
0.2767166514 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_054467_010 |
0.2788044011 |
- |
- |
- |