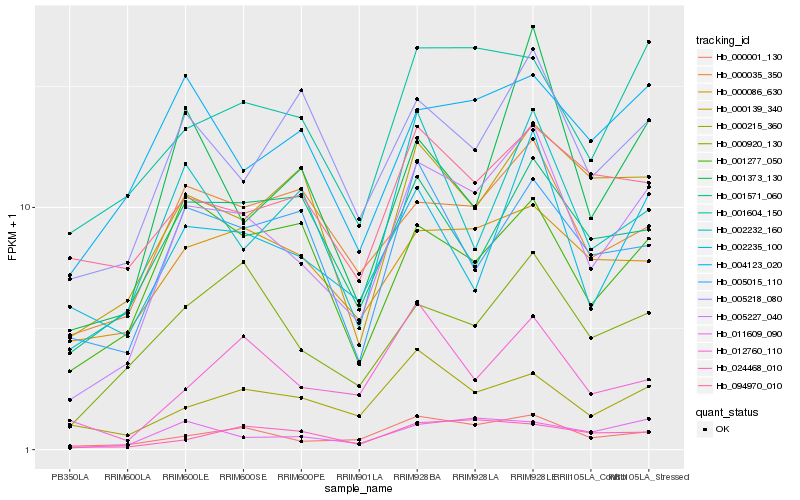
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005227_040 |
0.0 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000001_130 |
0.1164175017 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 3 |
Hb_001604_150 |
0.1551961591 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 4 |
Hb_011609_090 |
0.1567646719 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_002235_100 |
0.1585896142 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000920_130 |
0.1590883164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000035_350 |
0.1635662597 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005218_080 |
0.1668672156 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000139_340 |
0.167867477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000215_360 |
0.1705449838 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_002232_160 |
0.1713274867 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 12 |
Hb_000086_630 |
0.1725220013 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001373_130 |
0.1743781966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004123_020 |
0.1760324297 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001571_060 |
0.1768373726 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 16 |
Hb_001277_050 |
0.1779166323 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 17 |
Hb_024468_010 |
0.1795306083 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 18 |
Hb_005015_110 |
0.1799257858 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 19 |
Hb_094970_010 |
0.1800366374 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_012760_110 |
0.1813853907 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |