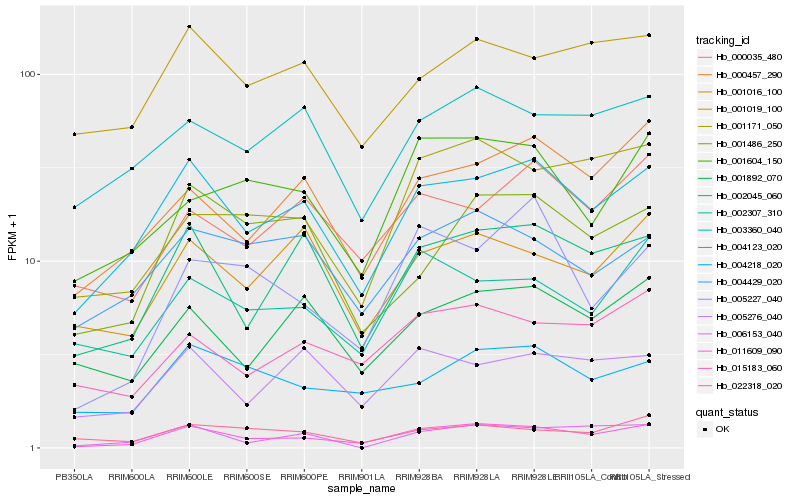
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_090 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_004123_020 |
0.105747915 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002045_060 |
0.1340433605 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 4 |
Hb_015183_060 |
0.1418282012 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 5 |
Hb_001171_050 |
0.1428209207 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 6 |
Hb_000457_290 |
0.1455063038 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 7 |
Hb_001604_150 |
0.1505931903 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |
| 8 |
Hb_002307_310 |
0.1507020227 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005227_040 |
0.1567646719 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_004218_020 |
0.1572730799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001892_070 |
0.1584482073 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 12 |
Hb_001016_100 |
0.1617976789 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 13 |
Hb_022318_020 |
0.1620556823 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 14 |
Hb_005276_040 |
0.1624602835 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 15 |
Hb_001019_100 |
0.1627132386 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 16 |
Hb_003360_040 |
0.16404843 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 17 |
Hb_004429_020 |
0.1643769954 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 18 |
Hb_000035_480 |
0.1652488423 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 19 |
Hb_001486_250 |
0.1655046075 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 20 |
Hb_006153_040 |
0.166307806 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |