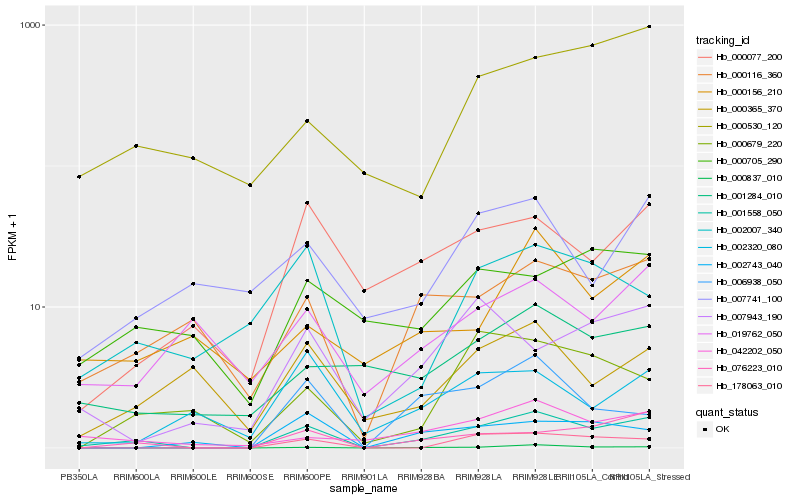
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001558_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641745 [Jatropha curcas] |
| 2 |
Hb_076223_010 |
0.2090067299 |
- |
- |
- |
| 3 |
Hb_000837_010 |
0.2372404098 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000116_360 |
0.2458502954 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 5 |
Hb_042202_050 |
0.2461005681 |
- |
- |
TPA: hypothetical protein ZEAMMB73_683745 [Zea mays] |
| 6 |
Hb_178063_010 |
0.2646853189 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 7 |
Hb_000077_200 |
0.2650841618 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 8 |
Hb_001284_010 |
0.2734164005 |
- |
- |
hypothetical protein CISIN_1g0279981mg, partial [Citrus sinensis] |
| 9 |
Hb_000679_220 |
0.2805219748 |
- |
- |
PREDICTED: uncharacterized protein LOC105637638 [Jatropha curcas] |
| 10 |
Hb_000530_120 |
0.2815206807 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 11 |
Hb_006938_050 |
0.2818353567 |
- |
- |
PREDICTED: uncharacterized protein LOC105648204 [Jatropha curcas] |
| 12 |
Hb_002007_340 |
0.2822413413 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000365_370 |
0.2826529561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007741_100 |
0.2859187588 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 15 |
Hb_002743_040 |
0.286938853 |
- |
- |
PREDICTED: uncharacterized protein LOC105785507 isoform X2 [Gossypium raimondii] |
| 16 |
Hb_019762_050 |
0.2877543884 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 17 |
Hb_007943_190 |
0.2877775716 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 18 |
Hb_000156_210 |
0.2885125902 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 19 |
Hb_000705_290 |
0.290736158 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 20 |
Hb_002320_080 |
0.2936772215 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |