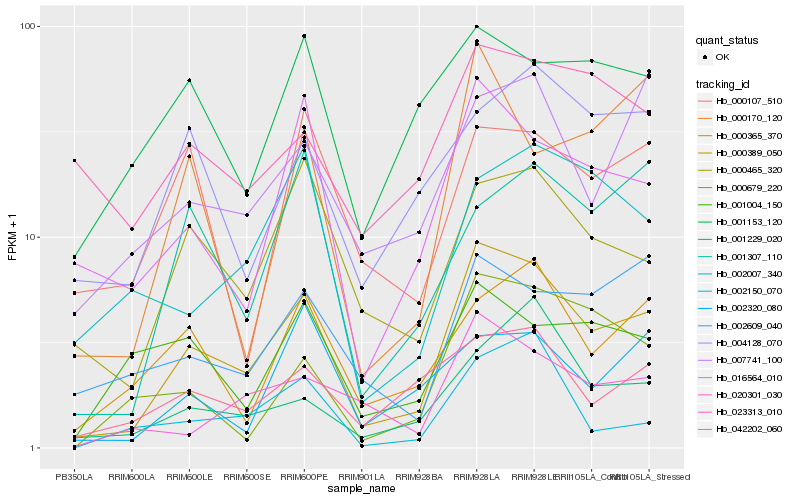
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000389_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 2 |
Hb_016564_010 |
0.1868542714 |
- |
- |
PREDICTED: uncharacterized protein LOC101764501 [Setaria italica] |
| 3 |
Hb_000465_320 |
0.1882546672 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 4 |
Hb_042202_060 |
0.1992247703 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 5 |
Hb_020301_030 |
0.2023407589 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 6 |
Hb_000365_370 |
0.2041332546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000679_220 |
0.209868802 |
- |
- |
PREDICTED: uncharacterized protein LOC105637638 [Jatropha curcas] |
| 8 |
Hb_007741_100 |
0.2107493082 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 9 |
Hb_002609_040 |
0.2107693612 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 10 |
Hb_001229_020 |
0.2109473907 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000170_120 |
0.2183380918 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_001307_110 |
0.218839817 |
- |
- |
PREDICTED: uncharacterized protein LOC105644507 [Jatropha curcas] |
| 13 |
Hb_004128_070 |
0.2196325528 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001004_150 |
0.2220043887 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 15 |
Hb_002007_340 |
0.2229197038 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002320_080 |
0.2243956139 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001153_120 |
0.2270178107 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 18 |
Hb_002150_070 |
0.227230694 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 19 |
Hb_000107_510 |
0.2284336281 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 20 |
Hb_023313_010 |
0.2288305599 |
- |
- |
PREDICTED: linolenate hydroperoxide lyase, chloroplastic [Jatropha curcas] |