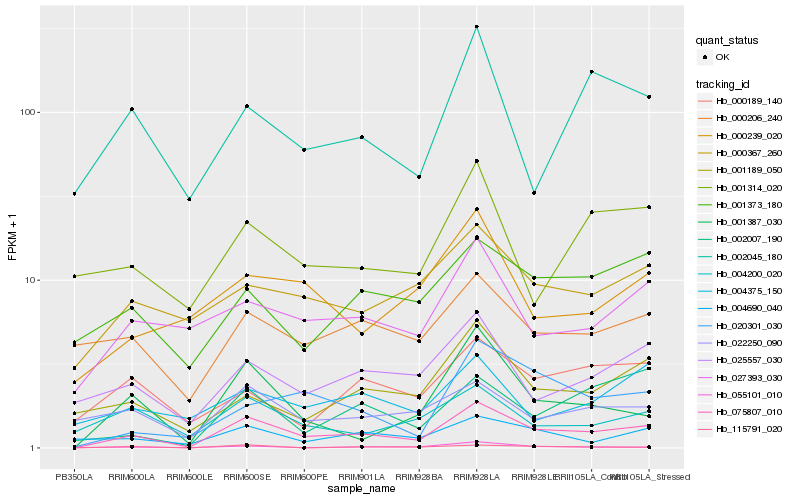
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075807_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631775 [Jatropha curcas] |
| 2 |
Hb_001387_030 |
0.2001803592 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 3 |
Hb_020301_030 |
0.2119593952 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_025557_030 |
0.2166707818 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 5 |
Hb_027393_030 |
0.2203351704 |
- |
- |
PREDICTED: uncharacterized protein At5g39865 [Jatropha curcas] |
| 6 |
Hb_002007_190 |
0.2234258722 |
- |
- |
hypothetical protein RCOM_1432220 [Ricinus communis] |
| 7 |
Hb_001189_050 |
0.2238896861 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 8 |
Hb_004690_040 |
0.2240950403 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 9 |
Hb_022250_090 |
0.2305599818 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 10 |
Hb_000367_260 |
0.2306708355 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 11 |
Hb_001373_180 |
0.2313600992 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 12 |
Hb_055101_010 |
0.2315782742 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_000206_240 |
0.2315860936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004200_020 |
0.234253814 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 15 |
Hb_002045_180 |
0.2343500669 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 16 |
Hb_000239_020 |
0.2372738369 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 17 |
Hb_115791_020 |
0.2380892183 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 18 |
Hb_004375_150 |
0.2397273235 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 19 |
Hb_001314_020 |
0.2397495055 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000189_140 |
0.2402429153 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |