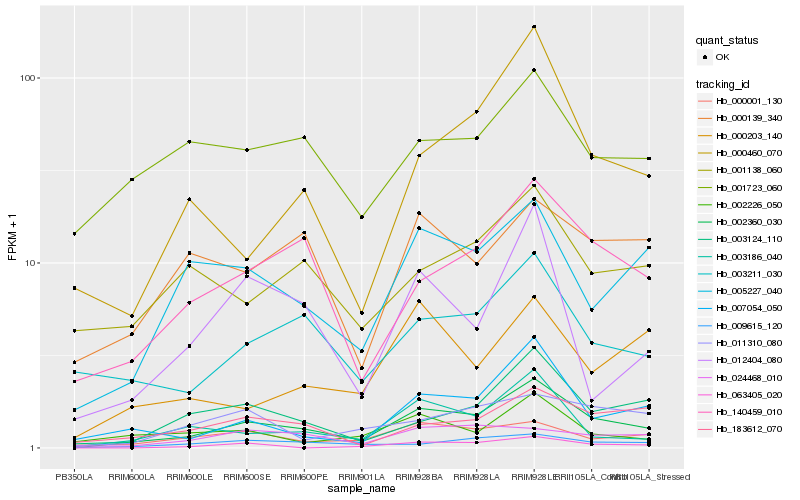
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002360_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 2 |
Hb_140459_010 |
0.1732532241 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 3 |
Hb_005227_040 |
0.1879397828 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_003124_110 |
0.191036556 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 5 |
Hb_007054_050 |
0.1927312476 |
- |
- |
- |
| 6 |
Hb_003186_040 |
0.1928697626 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 7 |
Hb_003211_030 |
0.1937478062 |
- |
- |
- |
| 8 |
Hb_009615_120 |
0.19660679 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 9 |
Hb_000001_130 |
0.1972359332 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 10 |
Hb_183612_070 |
0.2013704092 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000460_070 |
0.2054746046 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 12 |
Hb_024468_010 |
0.2125032443 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 13 |
Hb_063405_020 |
0.2136491188 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 14 |
Hb_011310_080 |
0.2146268773 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 15 |
Hb_000139_340 |
0.2180876302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001138_060 |
0.2186698383 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 17 |
Hb_000203_140 |
0.2196926519 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 18 |
Hb_001723_060 |
0.2215445153 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 19 |
Hb_002226_050 |
0.2216868165 |
- |
- |
PREDICTED: uncharacterized protein LOC105641366 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012404_080 |
0.2218919623 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |