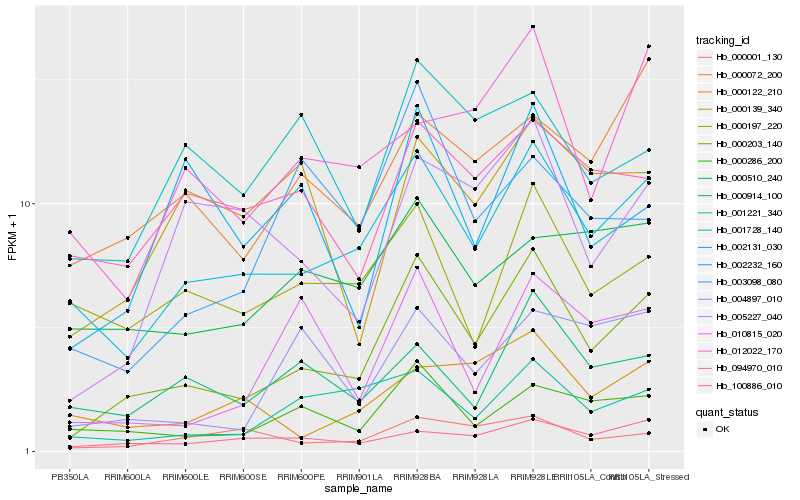
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_140 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 2 |
Hb_002131_030 |
0.139855104 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000286_200 |
0.1780958111 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_001221_340 |
0.1841451022 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 5 |
Hb_100886_010 |
0.1882466673 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_005227_040 |
0.1965886094 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_004897_010 |
0.1982172845 |
- |
- |
- |
| 8 |
Hb_000072_200 |
0.2005333792 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 9 |
Hb_010815_020 |
0.2011718895 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000914_100 |
0.201671616 |
- |
- |
unknown [Glycine max] |
| 11 |
Hb_012022_170 |
0.2067339685 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 12 |
Hb_000001_130 |
0.2070239134 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 13 |
Hb_000197_220 |
0.2086395434 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002232_160 |
0.2094703964 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 15 |
Hb_000510_240 |
0.2097403107 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_094970_010 |
0.2117925689 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000122_210 |
0.215688447 |
- |
- |
- |
| 18 |
Hb_001728_140 |
0.2162572596 |
- |
- |
- |
| 19 |
Hb_003098_080 |
0.2164517112 |
- |
- |
- |
| 20 |
Hb_000139_340 |
0.2166895153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |