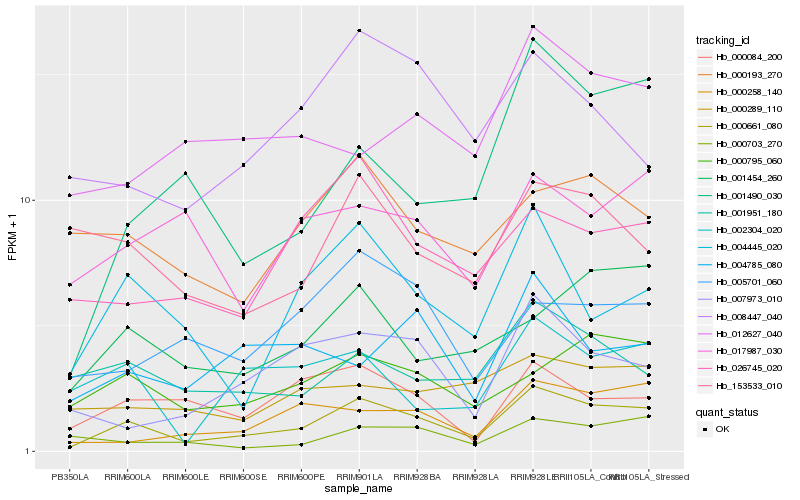
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_080 |
0.0 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 2 |
Hb_004445_020 |
0.1817874815 |
- |
- |
hypothetical protein JCGZ_13166 [Jatropha curcas] |
| 3 |
Hb_000795_060 |
0.183764301 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 4 |
Hb_000258_140 |
0.1892662629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001490_030 |
0.1914877711 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001454_260 |
0.1922193903 |
- |
- |
hypothetical protein RCOM_0681280 [Ricinus communis] |
| 7 |
Hb_000084_200 |
0.1966475356 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007973_010 |
0.1970574515 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 9 |
Hb_026745_020 |
0.200323788 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001951_180 |
0.2005706192 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_005701_060 |
0.2039658523 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 12 |
Hb_002304_020 |
0.2072558134 |
- |
- |
- |
| 13 |
Hb_000193_270 |
0.2079181319 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000289_110 |
0.2082155732 |
- |
- |
PREDICTED: cysteine synthase-like [Musa acuminata subsp. malaccensis] |
| 15 |
Hb_000703_270 |
0.2088294593 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 16 |
Hb_017987_030 |
0.2103697935 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 17 |
Hb_153533_010 |
0.210658366 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |
| 18 |
Hb_008447_040 |
0.2112083536 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 19 |
Hb_004785_080 |
0.2118774269 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 20 |
Hb_012627_040 |
0.2127685771 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |