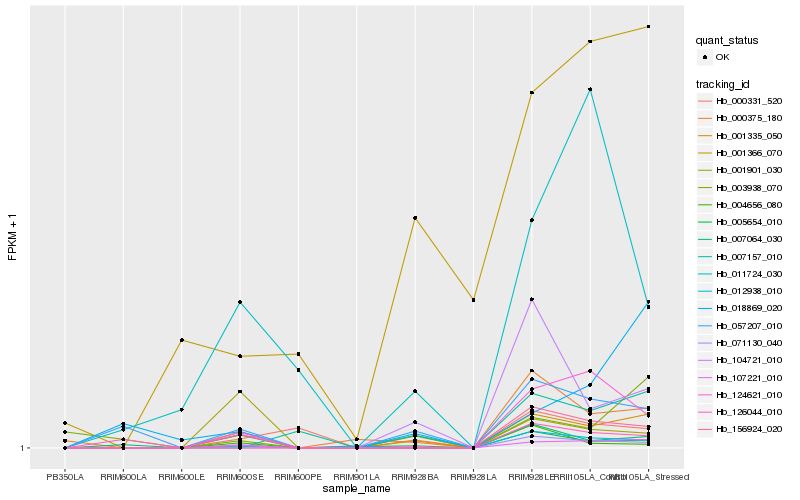
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001335_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_107221_010 |
0.1131196356 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_005654_010 |
0.2116970778 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_071130_040 |
0.2232982616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_057207_010 |
0.2594781269 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 6 |
Hb_000375_180 |
0.2619457158 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 7 |
Hb_007064_030 |
0.2902068061 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 8 |
Hb_124621_010 |
0.290749999 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 9 |
Hb_012938_010 |
0.3057401383 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_001901_030 |
0.3220373211 |
- |
- |
- |
| 11 |
Hb_126044_010 |
0.3241028024 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 12 |
Hb_000331_520 |
0.3281608826 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 13 |
Hb_001366_070 |
0.337973727 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_104721_010 |
0.3384264711 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 15 |
Hb_007157_010 |
0.3406635038 |
- |
- |
- |
| 16 |
Hb_004656_080 |
0.3428307579 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_011724_030 |
0.3433258023 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 18 |
Hb_003938_070 |
0.3476985355 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_018869_020 |
0.3508555762 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 20 |
Hb_156924_020 |
0.3525952782 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |