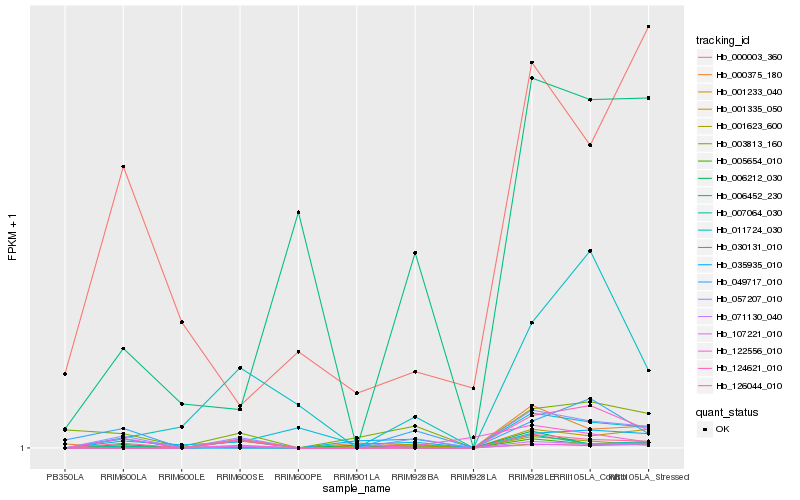
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_057207_010 |
0.0 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 2 |
Hb_126044_010 |
0.2280137441 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_001335_050 |
0.2594781269 |
- |
- |
- |
| 4 |
Hb_107221_010 |
0.2770455755 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_005654_010 |
0.2805895693 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_007064_030 |
0.2838911764 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_001623_600 |
0.284247163 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 8 |
Hb_001233_040 |
0.2904983457 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 9 |
Hb_003813_160 |
0.2956856332 |
- |
- |
- |
| 10 |
Hb_071130_040 |
0.3057842275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000375_180 |
0.3253831245 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 12 |
Hb_122556_010 |
0.3255062551 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 13 |
Hb_006452_230 |
0.3261518443 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 14 |
Hb_035935_010 |
0.3281056242 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 15 |
Hb_124621_010 |
0.3308206395 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 16 |
Hb_011724_030 |
0.3310188152 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 17 |
Hb_049717_010 |
0.3350630048 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 18 |
Hb_000003_360 |
0.3415965996 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 19 |
Hb_006212_030 |
0.3439683851 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 20 |
Hb_030131_010 |
0.3443118862 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |