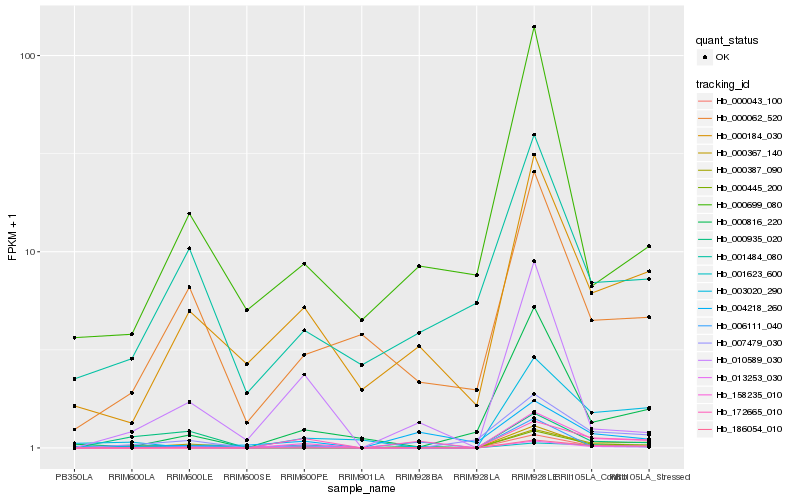
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 2 |
Hb_000387_090 |
0.2276288196 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 3 |
Hb_013253_030 |
0.2349048966 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000184_030 |
0.2452795853 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007479_030 |
0.2498196075 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 6 |
Hb_000367_140 |
0.2539887862 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 7 |
Hb_001623_600 |
0.2568669289 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 8 |
Hb_000043_100 |
0.2651326344 |
- |
- |
PREDICTED: uncharacterized protein LOC105797583 [Gossypium raimondii] |
| 9 |
Hb_158235_010 |
0.2664627347 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 10 |
Hb_000062_520 |
0.275929143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004218_260 |
0.2779498457 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 12 |
Hb_003020_290 |
0.281163728 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 13 |
Hb_010589_030 |
0.2817074964 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 14 |
Hb_000816_220 |
0.2822225927 |
- |
- |
PREDICTED: CBL-interacting protein kinase 18-like [Jatropha curcas] |
| 15 |
Hb_186054_010 |
0.28225294 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_006111_040 |
0.2834665512 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 17 |
Hb_001484_080 |
0.2857825437 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 18 |
Hb_000699_080 |
0.2928156181 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_172665_010 |
0.2945922632 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_000935_020 |
0.2986643788 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |