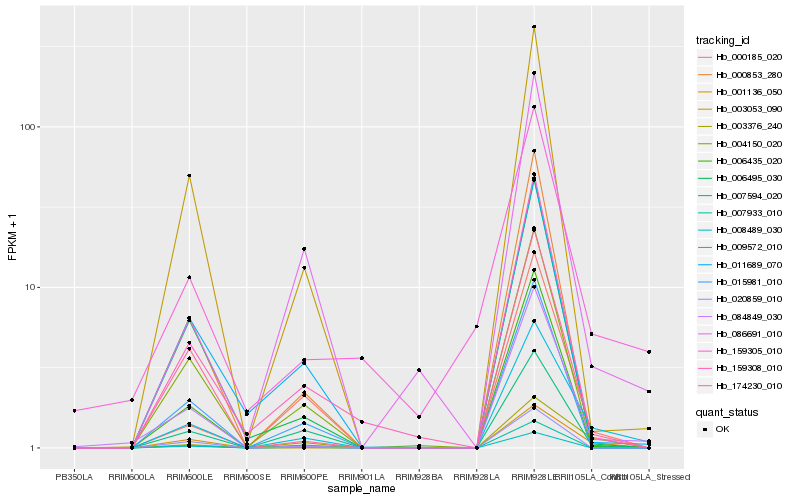
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009572_010 |
0.0 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 2 |
Hb_003376_240 |
0.1508842199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_084849_030 |
0.1574316113 |
- |
- |
PREDICTED: uncharacterized protein LOC105634080 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001136_050 |
0.1633901471 |
- |
- |
- |
| 5 |
Hb_007594_020 |
0.1742017236 |
- |
- |
hypothetical protein JCGZ_18350 [Jatropha curcas] |
| 6 |
Hb_000185_020 |
0.1764880289 |
- |
- |
- |
| 7 |
Hb_174230_010 |
0.1810057008 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 8 |
Hb_003053_090 |
0.1826344848 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_004150_020 |
0.1846334089 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 10 |
Hb_086691_010 |
0.1877550518 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 11 |
Hb_159308_010 |
0.1879972932 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 12 |
Hb_000853_280 |
0.1895613681 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 13 |
Hb_015981_010 |
0.1898020003 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_159305_010 |
0.1908566032 |
- |
- |
rpl2 [Jatropha curcas] |
| 15 |
Hb_007933_010 |
0.1914379045 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 16 |
Hb_006495_030 |
0.1921988405 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 17 |
Hb_011689_070 |
0.193630465 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006435_020 |
0.1936426694 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_020859_010 |
0.1941950734 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X6 [Populus euphratica] |
| 20 |
Hb_008489_030 |
0.1945471246 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |