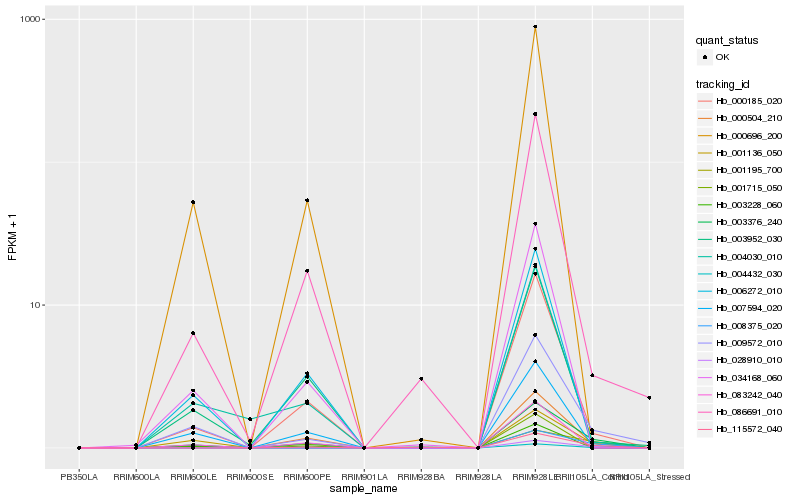
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009572_010 |
0.1508842199 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 3 |
Hb_000185_020 |
0.1810557536 |
- |
- |
- |
| 4 |
Hb_000504_210 |
0.1849342709 |
- |
- |
hypothetical protein JCGZ_10251 [Jatropha curcas] |
| 5 |
Hb_115572_040 |
0.195532347 |
- |
- |
PREDICTED: uncharacterized protein LOC105632710 [Jatropha curcas] |
| 6 |
Hb_007594_020 |
0.205567938 |
- |
- |
hypothetical protein JCGZ_18350 [Jatropha curcas] |
| 7 |
Hb_086691_010 |
0.2162064994 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 8 |
Hb_028910_010 |
0.2206095779 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_001136_050 |
0.2216505914 |
- |
- |
- |
| 10 |
Hb_004432_030 |
0.2245406284 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 11 |
Hb_003952_030 |
0.2256320292 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 12 |
Hb_001195_700 |
0.2282080682 |
- |
- |
- |
| 13 |
Hb_008375_020 |
0.2289889704 |
- |
- |
PREDICTED: uncharacterized protein LOC104894372 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_083242_040 |
0.2395837063 |
- |
- |
- |
| 15 |
Hb_001715_050 |
0.2399616365 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase AOP1 [Jatropha curcas] |
| 16 |
Hb_003228_060 |
0.2399778659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000696_200 |
0.2413746731 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 18 |
Hb_006272_010 |
0.2422495141 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 19 |
Hb_004030_010 |
0.2428751208 |
- |
- |
hypothetical protein CICLE_v10013050mg [Citrus clementina] |
| 20 |
Hb_034168_060 |
0.2437623879 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |