| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
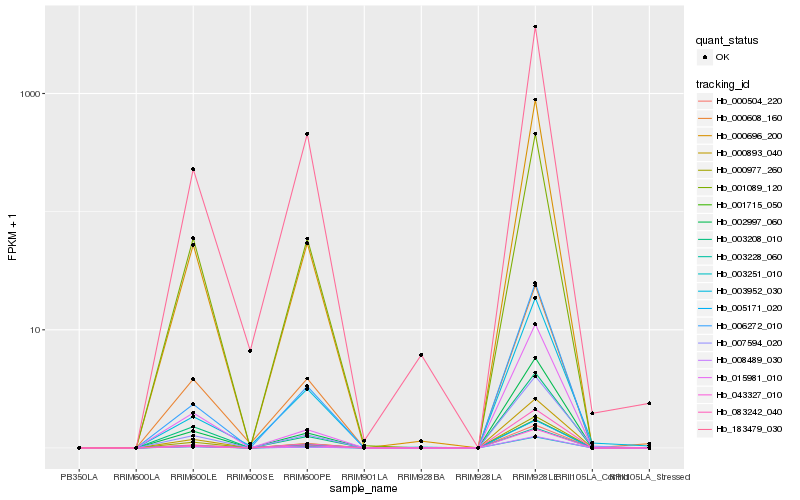
Hb_007594_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_18350 [Jatropha curcas] |
| 2 |
Hb_043327_010 |
0.0723106451 |
- |
- |
PREDICTED: uncharacterized protein LOC105785093, partial [Gossypium raimondii] |
| 3 |
Hb_005171_020 |
0.0752446515 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 4 |
Hb_002997_060 |
0.0766149137 |
- |
- |
hypothetical protein POPTR_0011s10730g [Populus trichocarpa] |
| 5 |
Hb_006272_010 |
0.0842221503 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 6 |
Hb_000504_220 |
0.0842807765 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 7 |
Hb_001089_120 |
0.0882721733 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 8 |
Hb_003952_030 |
0.0894439462 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 9 |
Hb_003251_010 |
0.0895528612 |
- |
- |
hypothetical protein MIMGU_mgv1a025126mg [Erythranthe guttata] |
| 10 |
Hb_008489_030 |
0.0900492497 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 11 |
Hb_183479_030 |
0.0910865572 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001715_050 |
0.0913920266 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase AOP1 [Jatropha curcas] |
| 13 |
Hb_000893_040 |
0.0921472128 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 14 |
Hb_003228_060 |
0.0939431826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000696_200 |
0.0944254369 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 16 |
Hb_083242_040 |
0.0973485773 |
- |
- |
- |
| 17 |
Hb_000977_260 |
0.1003323413 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 18 |
Hb_000608_160 |
0.1019929922 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 19 |
Hb_003208_010 |
0.1019967883 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_015981_010 |
0.1049345788 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |