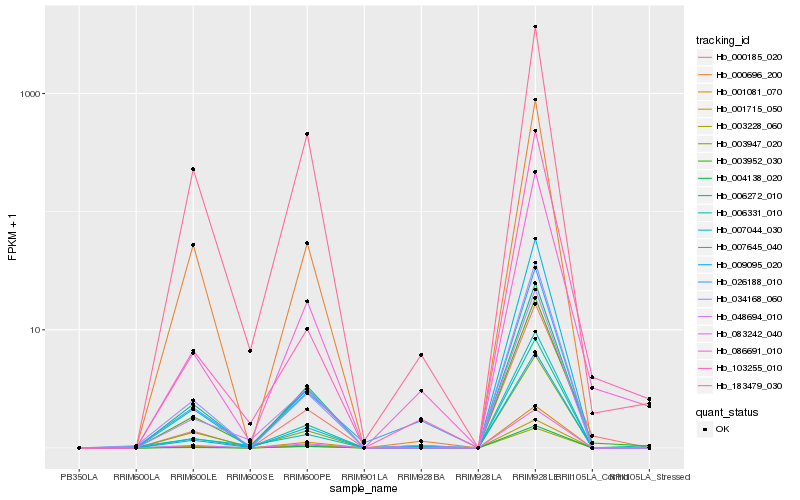
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086691_010 |
0.0 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 2 |
Hb_000185_020 |
0.0874044991 |
- |
- |
- |
| 3 |
Hb_009095_020 |
0.0881204544 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 4 |
Hb_003952_030 |
0.0961651535 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 5 |
Hb_026188_010 |
0.108312523 |
- |
- |
PREDICTED: tropinone reductase homolog [Jatropha curcas] |
| 6 |
Hb_006331_010 |
0.1093218116 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 7 |
Hb_001081_070 |
0.1137035851 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 8 |
Hb_034168_060 |
0.1138328741 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |
| 9 |
Hb_003947_020 |
0.1143975508 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 10 |
Hb_004138_020 |
0.114809437 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 11 |
Hb_083242_040 |
0.1169285464 |
- |
- |
- |
| 12 |
Hb_007044_030 |
0.1187912906 |
- |
- |
Os06g0273400 [Oryza sativa Japonica Group] |
| 13 |
Hb_001715_050 |
0.1197214328 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase AOP1 [Jatropha curcas] |
| 14 |
Hb_007645_040 |
0.1218538934 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |
| 15 |
Hb_003228_060 |
0.1226328123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_103255_010 |
0.1249171737 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 17 |
Hb_048694_010 |
0.1250849443 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_183479_030 |
0.1260306017 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_006272_010 |
0.1269644116 |
- |
- |
PREDICTED: cytochrome P450 86B1 [Vitis vinifera] |
| 20 |
Hb_000696_200 |
0.1272009363 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |