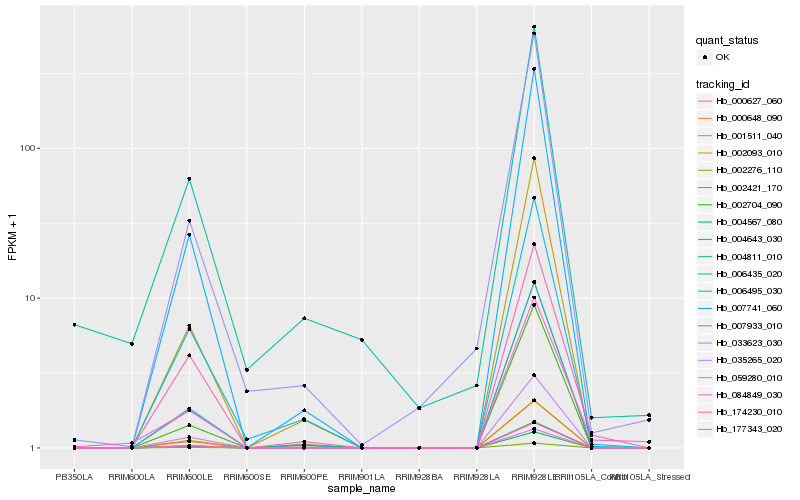
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084849_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634080 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006435_020 |
0.1168875768 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_174230_010 |
0.1215319967 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 4 |
Hb_007741_060 |
0.1277930902 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 5 |
Hb_035265_020 |
0.1297515186 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_177343_020 |
0.1302189361 |
- |
- |
PREDICTED: uncharacterized protein LOC104751876 [Camelina sativa] |
| 7 |
Hb_000648_090 |
0.1304746582 |
- |
- |
- |
| 8 |
Hb_000627_060 |
0.1305711934 |
- |
- |
- |
| 9 |
Hb_004643_030 |
0.1309365657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004567_080 |
0.1309450391 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 11 |
Hb_002421_170 |
0.1317113318 |
- |
- |
hypothetical protein POPTR_0003s18240g [Populus trichocarpa] |
| 12 |
Hb_004811_010 |
0.1328828248 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 13 |
Hb_002093_010 |
0.1338157256 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 14 |
Hb_001511_040 |
0.1345866839 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 15 |
Hb_006495_030 |
0.1384559324 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 16 |
Hb_002276_110 |
0.1388054182 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_033623_030 |
0.1398213172 |
- |
- |
PREDICTED: isoprene synthase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_059280_010 |
0.1398430162 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 19 |
Hb_002704_090 |
0.1411649494 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_007933_010 |
0.1421641368 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |